| Issue |
Parasite
Volume 31, 2024
|
|
|---|---|---|
| Article Number | 48 | |
| Number of page(s) | 14 | |
| DOI | https://doi.org/10.1051/parasite/2024046 | |
| Published online | 14 August 2024 | |
urn:lsid:zoobank.org:pub:B55402B8-4513-4899-ACFB-FD5D8746C46D
Research Article
Morphology, complete mitochondrial genome, and molecular phylogeny of Rhabdias macrocephalum n. sp. (Nematoda: Rhabdiasidae) from Diploderma splendidum (Reptilia: Agamidae)
Morphologie, génome mitochondrial complet et phylogénie moléculaire de Rhabdias macrocephalum n. sp. (Nematoda : Rhabdiasidae) de Diploderma splendidum (Reptilia : Agamidae)
1
Hebei Collaborative Innovation Center for Eco‐Environment, Hebei Key Laboratory of Animal Physiology, Biochemistry and Molecular Biology, College of Life Sciences, Hebei Normal University, 050024 Shijiazhuang, Hebei Province, PR China
2
Hebei Research Center of the Basic Discipline Cell Biology, Ministry of Education Key Laboratory of Molecular and Cellular Biology, 050024 Shijiazhuang, Hebei Province, PR China
* Corresponding author: liangliangex369@126.com
Received:
3
April
2024
Accepted:
19
July
2024
Species of the genus Rhabdias Stiles & Hassall, 1905 are common parasitic nematodes occurring in the lungs of amphibians and reptiles worldwide. In the present study, Rhabdias macrocephalum n. sp. is described using integrated morphological methods (light and scanning electron microscopy) and molecular approaches (sequencing of the nuclear 28S and ITS regions, and mitochondrial cox1, cox2, and 12S genes) based on specimens collected from the green striped tree dragon Diploderma splendidum (Barbour & Dunn) (Reptilia: Agamidae) in China. The complete mitochondrial genome of R. macrocephalum n. sp. was sequenced and annotated: it is 14,819 bp in length, including 12 protein coding genes (missing atp8), 22 tRNA genes, 2 rRNA genes and three non-coding regions. The gene arrangement of R. macrocephalum n. sp. is different from all of the currently available mitogenomes of nematodes and represents a novel type of mitochondrial gene arrangement reported in Nematoda. Molecular phylogenetic results based on the ITS + 28S data support the monophyly of Entomelas, Pneumonema, Serpentirhabdias, and Rhabdias, and showed R. macrocephalum n. sp. forming a most basal lineage in Rhabdias.
Résumé
Les espèces du genre Rhabdias Stiles & Hassall, 1905 sont des nématodes parasites courants présents dans les poumons des amphibiens et des reptiles du monde entier. Dans cette étude, Rhabdias macrocephalum n. sp. est décrit à l’aide de méthodes morphologiques intégrées (microscopie optique et électronique à balayage) et d’approches moléculaires (séquençage des régions nucléaires 28S et ITS et des gènes mitochondriaux cox1, cox2 et 12S) basées sur des spécimens collectés chez le lézard Diploderma splendidum (Barbour & Dunn) (Reptilia : Agamidae) de Chine. Le génome mitochondrial complet de R. macrocephalum n. sp. a été séquencé et annoté : il a une longueur de 14 819 pb, dont 12 gènes codants pour des protéines (atp8 manquant), 22 gènes d’ARNt, 2 gènes d’ARNr et trois régions non codantes. L’arrangement génétique de R. macrocephalum n. sp. est différent de tous les mitogénomes de nématodes actuellement disponibles et représente un nouveau type d’arrangement de gènes mitochondriaux signalé chez les nématodes. Les résultats phylogénétiques moléculaires basés sur les données ITS + 28S ont soutenu la monophylie d’Entomelas, Pneumonema, Serpentirhabdias et Rhabdias, et ont montré que R. macrocephalum n. sp. forme la lignée la plus basale chez Rhabdias.
Key words: Zooparasitic nematodes / Rhabdiasidae / Integrative taxonomy / Mitochondrial genome / Phylogeny
© J. Zeng et al., published by EDP Sciences, 2024
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The genus Rhabdias (Nematoda: Rhabditida) is the largest group in the family Rhabdiasidae, and currently comprises over 90 nominal species mainly parasitic in the lungs of amphibians and reptiles worldwide [15, 16, 22, 47]. To date, a total of 8 species of Rhabdias have been reported in China, namely R. bicornis Lu, 1934, R. incerta Wilkie, 1930, R. brevicauda Chu, 1936, R. nipponica Yamaguti, 1935, R. bufonis (Schrank, 1788), R. globocephala Kung & Wu, 1945, R. japalurae Kuzmin, 2003, and R. kafunata Sata, Takeuchi & Nakano, 2020 [14, 21, 29, 43, 51, 52]. However, our present knowledge of the species composition of Rhabdias nematodes in China is still far from complete.
It is not easy to precisely identify specimens of Rhabdias to species level based only on morphological characters, due usually to a lack of males and the extraordinary morphological similarity in females. Recently, some genetic data [i.e., large nuclear ribosomal DNA (28S), internal transcribed spacer (ITS), mitochondrial cytochrome c oxidase subunit 1 (cox1), and 12S small subunit ribosomal RNA gene] and mitochondrial genomes have been successfully used to identify species, discover sibling or cryptic species, and evaluate evolutionary relationships of Rhabdiasidae [1, 15, 16, 28, 31, 36, 46, 47, 52]. However, the current genetic database, especially the mitogenomes for the rhabdiasid nematodes, remains very insufficient. To date, only R. bufonis and R. kafunata have been reported for the complete mitochondrial genomes in the Rhabdiasidae [28, 52].
In the present study, a new species of Rhabdias collected from the green striped tree dragon Diploderma splendidum (Barbour & Dunn) (Reptilia: Agamidae) in China was precisely identified using integrated morphological methods (light and scanning electron microscopy) and molecular approaches (sequencing of the nuclear 28S and ITS regions and mitochondrial cox1, cox2, and 12S genes). Additionally, in order to enrich the mitogenomic data and reveal the patterns of mitogenomic evolution of the Rhabdiasidae, the complete mitochondrial genome of this new species was sequenced and annotated. Moreover, in order to determine the phylogenetic position of this new species within Rhabdias, phylogenetic analyses were performed based on the 28S + ITS sequences, using maximum likelihood (ML) and Bayesian inference (BI), respectively.
Materials and methods
Morphological observation
In 2021, a total of 26 nematode specimens of Rhabdias were sent to the author’s (Li L.) laboratory for species identification, which were recovered from the lung of a dead green striped tree dragon D. splendidum by a local veterinarian in Qinzhou, Guangxi Zhuang Autonomous Region, China. Specimens were fixed and stored in 80% ethanol until the morphological study. For light microscopy, nematode specimens were cleared in 50% glycerin, then examined and photographed using a Nikon® optical microscope (Nikon ECLIPSE Ni-U, Nikon Corporation, Tokyo, Japan). For scanning electron microscopy (SEM), the anterior and posterior ends of specimens were transferred to 4% formaldehyde solution, then post-fixed in 1% OsO4, dehydrated via an ethanol series and acetone and critical point dried. The specimens were coated with gold and examined using a Hitachi S-4800 scanning electron microscope (Hitachi Ltd., Tokyo, Japan) at an accelerating voltage of 20 kV. All measurements in the text are in micrometers unless otherwise stated. Type specimens were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, and the National Zoological Museum, Beijing, China.
Molecular procedures
A total of three female specimens were randomly selected for the molecular analysis. Genomic DNA from each individual was extracted using a Column Genomic DNA Isolation Kit (Shanghai Sangon, Shanghai, China), according to the manufacturer’s instructions. DNA was eluted in elution buffer and kept at −20 °C until use. The primers and cycling conditions for amplifying different target regions by polymerase chain reaction (PCR) are provided in Table 1. All PCR reactions were performed in 50 μL consisting of 10 mM Tris HCl at pH 8.4, 50 mM KCl, 3.0 mM MgCl2, 250 μM of each dNTP, 50 pmol of each primer, and 1.5 U of Taq polymerase (Takara Bio Inc., Kusatsu, Shiga, Japan) in a thermocycler (model 2720; Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA).
The primers and cycling conditions for amplifying different target regions by polymerase chain reaction (PCR) in the present study.
PCR products were checked on GoldView-stained 1.5% agarose gel and purified by the Column PCR Product Purification Kit (Shanghai Sangon). Sequencing for each sample was carried out for both strands using a DyeDeoxyTerminator Cycle Sequencing Kit v.2 (Applied Biosystems). The 28S, ITS, cox1, cox2, and 12S sequences obtained herein were deposited in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov).
Mitochondrial genome sequencing, assembly, and annotation
A total of 30 Gb clean genomic data were generated using the Pair-End 150 sequencing method on the Illumina NovaSeq 6000 platform by Novogene (Tianjin, China). The complete mitochondrial genomes were assembled using GetOrganelle v1.7.2a [12]. Protein coding genes (PCGs), rRNAs, and tRNAs were annotated using MitoS web server (http://mitos2.bioinf.uni-leipzig.de/index.py) and MitoZ v2.4 [33]. The open reading frame (ORF) of each PCG was confirmed manually by the web version of ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The “lost” tRNA genes ignored by both MitoS and MitoZ, were identified using BLAST based on a database of the existing tRNA sequences of nematodes. The secondary structures of tRNAs were predicted by ViennaRNA module [9], building on MitoS2 [2] and RNAstructure v6.3 [40], followed by manual correction. MitoZ v2.4 was used to visualize and depict gene element features [33]. The base composition, amino acid usage, and relative synonymous codon usage (RSCU) were calculated by Python script, which refers to Codon Adaptation Index (CAI) [23]. The total length of the base composition included ambiguous bases. The base skew analysis was used to describe the base composition of nucleotide sequences. The complete mitochondrial genome of this new species obtained was deposited in the NCBI database (http://www.ncbi.nlm.nih.gov).
Phylogenetic analyses
Phylogenetic analyses of rhabdiasid nematodes were performed based on the ITS + 28S sequences using maximum likelihood (ML) with IQ-TREE [34] and Bayesian inference (BI) with MrBayes [41]. Caenorhabditis elegans (Rhabditida: Rhabditidae) was chosen as the out-group. The in-group included 46 rhabdiasid species representing six genera. Detailed information on species included in the phylogenetic analyses is provided in Table 2. Genes were aligned separately using the MAFFT v7.313 multiple sequence alignment program under the iterative refinement method of E-INS-I [13]. In addition, partially ambiguous bases were manually inspected and removed. The aligned and pruned sequences were concatenated into a matrix by PhyloSuite v1.2.2. The TVM + F+I + I+R2 model was selected for ML analyses. The GTR + F+G4 models were selected for BI analyses. Reliabilities for ML inference were tested using 1000 bootstrap replications, and BIC analysis was run for 5 × 106 MCMC generations.
Detailed information on the representatives of Rhabdiasidae with their genetic data included in the phylogenetic analyses.
Results
Description of Rhabdias macrocephalum n. sp. (Figs. 1–3)
urn:lsid:zoobank.org:act:BB1898CA-CD19-4568-9A43-84E2AD6FF185
 |
Figure 1 Photomicrographs of Rhabdias macrocephalum n. sp. from Diploderma splendidum in China. A: entire body (vulva arrowed), lateral view; B: anterior part of body, lateral view; C: region of vulva, lateral view; D: cephalic extremity, lateral view; E: posterior part of body (cloaca arrowed), lateral view. |
Type host: Green striped tree dragon Diploderma splendidum (Barbour & Dunn) (Reptilia: Agamidae).
Type locality: Qinzhou City, Guangxi Zhuang Autonomous Region, China.
Site in host: Lung.
Type specimens: Holotype: 1 female (HBNU–N–R20240315ZL); paratypes: 22 females (HBNU–N–R20240316ZL), deposited in the College of Life Sciences, Hebei Normal University, Hebei Province; 3 females (NZMC–PN_144–146), deposited in the National Zoological Museum, Beijing, China.
Etymology: The specific name refers to the inflated cephalic end of the present specimens.
GenBank accession: PP544391–PP544393 (28S), PP544389–PP544390 (ITS), PP533065–PP533067 (cox1), PP544387–PP544388 (12S), PP550091 (cox2), PP874272 (mitogenome).
Diagnosis: Body relatively large, gradually tapering from mid-region towards anterior and posterior ends (Fig. 1A). Cephalic extremity conspicuously inflated to form cephalic bulb (Figs. 1B, D, 2A, C, 3A). Cuticle slightly or inconspicuously inflated at anterior region of body, then distinctly inflated to form irregular folds from more or less posterior region of nerve ring (Figs. 1B, 2A), and conspicuously inflated at vulval and caudal region (Figs. 1C, E, 2B, E). Esophagus club-shaped, possessing an indistinct dilation at anterior region of nerve ring, posterior end distinctly expanded to esophageal bulb (Figs. 1B, 2A). Excretory pore just posterior to nerve ring (Figs. 2A, 3E). Tail conical, sharply pointed, abruptly tapering from anus posteriorly, gradually tapering from approximately 1/2 of tail (Figs. 1A, E, 2B, 3B).
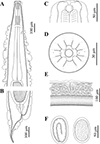 |
Figure 2 Line drawings of Rhabdias macrocephalum n. sp. from Diploderma splendidum in China. A: anterior part of body, lateral view; B: posterior part of body, lateral view; C: cephalic extremity, lateral view; D: cephalic extremity, apical view; E: region of vulva, lateral view; F: eggs. |
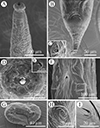 |
Figure 3 Scanning electron micrographs of Rhabdias macrocephalum n. sp. from Diploderma splendidum in China. A: anterior part of body, lateral view; B: tail, ventral view; C: magnified image of lateral cuticular pore; D: cephalic extremity (single papilla on each lip arrowed), apical view; E: magnified image of amphid; F: mid-body at level of vulva, sublateral view; G: egg with developed larva; H: magnified image of lateral cuticular pores on the tail; I: magnified image of lateral cuticular pores on the middle of body. Abbreviations: sl, submedian lip; ll, lateral lip. |
General (Based on 10 gravid individuals): Body 14.0–18.0 (17.0) mm long, maximum width 976–1293 (1112). Cuticular pores arranged laterally into 2 longitudinal rows along entire body (Fig. 3B, H, I). Oral opening simple, nearly rounded, surrounded by six small lips (two lateral and four submedian) reduced to elongated elevations (Figs. 2D, 3D); submedian lips located closer to edge of oral opening than lateral lips, each lip bearing single papilla (Figs. 2D, 3D). Small amphids located at base of lateral lip (Figs. 2D, 3E). Vestibulum narrow, cylindrical, cuticularized. Buccal capsule small, cup-like, with well sclerotized walls, 20.0–22.5 (21.5) deep, 25.0–30.0 (26.8) wide (Figs. 1D, 2A, C). Esophagus 870–980 (937) in total length, representing 5.35–6.00 (5.67) % of body length. Nerve ring 251–319 (278) from cephalic extremity. Uteri didelphic and amphidelphic, typical of Rhabdias; vulval opening with slightly protruding lips, 8.29–9.76 (9.35) mm from cephalic extremity, representing 53.6–58.5 (56.6) % of body length (Figs. 1A, C, 2E, 3F). Uteri thin-walled, filled with well developed, embryonated or unembryonated eggs (Figs. 1C, 2E, F, 3G). Eggs oval, with smooth thin-shell, 72–116 (91) × 34–68 (49) (n = 20). Tail 261–328 (302) long, representing 1.52–2.02 (1.83) % body length.
Genetic characterization
Three partial 28S sequences of R. macrocephalum n. sp. obtained here are all 551 bp, with no nucleotide divergence detected. Pairwise comparison of the partial 28S sequences of R. macrocephalum n. sp. obtained here with that of Rhabdias available in GenBank, displayed 1.45% (R. pseudosphaerocephala, MH516124; R. breviensis, MH516101) to 3.58% (R. tarichae, MH023521) nucleotide divergence. Two partial ITS sequences of R. macrocephalum n. sp. obtained here are both 700 bp, with no nucleotide divergence detected. Pairwise comparison of the partial ITS sequences of R. macrocephalum n. sp. obtained here with that of Rhabdias spp. available in GenBank, displayed 7.80% (R. breviensis, MH516064) to 16.8% (R. stomatica, MW522544) nucleotide divergence. Three partial cox1 sequences of R. macrocephalum n. sp. obtained here are all 655 bp, with no nucleotide divergence detected. Pairwise comparison of the partial cox1 sequences of R. macrocephalum n. sp. obtained here with that of Rhabdias spp. available in GenBank, displayed 8.70% (R. nipponica, LC671281) to 15.4% (R. lamothei, KC130747) nucleotide divergence. Two partial 12S sequences of R. macrocephalum n. sp. obtained here are both 474 bp, with no nucleotide divergence detected. Pairwise comparison of the partial 12S sequences of R. macrocephalum n. sp. obtained here with that of Rhabdias spp. available in GenBank, displayed 8.91% (R. engelbrechti, MG428408) to 11.5% (R. mariauxi, FN395318) nucleotide divergence. One partial cox2 sequence of R. macrocephalum n. sp. obtained here is 554 bp. Pairwise comparison of the partial cox2 sequence of R. macrocephalum n. sp. obtained here with that of Rhabdias spp. available in GenBank, displayed 11.2% (R. bufonis) to 13.2% (R. kafunata) nucleotide divergence.
Characterization of complete mitogenome
The mitogenome of R. macrocephalum n. sp. had 14,819 bp, containing 36 genes, including 12 PCGs (missing atp8) (cox1–3, cytb, nad1–6, nad4L and atp6), 22 tRNA genes, and 2 rRNA genes (rrnL and rrnS) (Fig. 4, Table 3). All genes were transcribed from the same DNA strand. There were three non-coding regions in the mitogenome of R. macrocephalum n. sp. (NCR1 is 441 bp, between nad5 and tRNA-Ala; NCR2 is 421 bp, between tRNA-Ala and tRNA-Met; NCR3 is 439 bp, between tRNA-Met and tRNA-Cys) (Fig. 4). The nucleotide contents of mitogenome of R. macrocephalum n. sp. are provided in Table 4. The overall A + T contents in the mitogenome of R. macrocephalum n. sp. was 77.5%, showing a strong nucleotide compositional bias toward A + T (Table 4).
 |
Figure 4 Gene maps of the mitochondrial genomes of Rhabdias macrocephalum n. sp. Abbreviations: NCR, non-coding region; PCG, protein coding gene; rRNA, ribosomal RNA; tRNA, transfer RNA. |
Annotations and gene organization of Rhabdias macrocephalum n. sp. Positive number in the “Gap or overlap” column indicates the length of intergenic sequence, and the negative number indicates the length (absolute number) that adjacent genes overlap (negative sign). The forward strand is marked as “+” and the reverse strand as “−”.
Base composition and skewness of Rhabdias macrocephalum n. sp.
The 12 PCGs of the mitogenome of R. macrocephalum n. sp. had 10,377 bp (excluding termination codons), and ranged in size from 231 bp (nad4L) to 1623 bp (cox1), which encoded 3448 amino acids. Among the 12 PCGs of R. macrocephalum n. sp., six genes (cox1, nad1, nad2, cytb, cox3, and nad4) used TTG as the start codon, followed by ATT for five genes (nad3, cox2, nad5, nad4L, and atp6), and ATG was used by nad6. TAA was the most commonly used termination codon (cox1, cox2, nad5, nad6, nad1, atp6, nad2, and cytb), and four genes including nad3, nad4L, cox3, and nad4 used TAG (Table 3). The component and usages of codons in the mitogenome of R. macrocephalum n. sp. are shown in Figure 5. The lengths of 22 tRNAs of R. macrocephalum n. sp. are provided (Table 3).
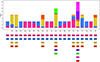 |
Figure 5 Relative synonymous codon usage (RSCU) of Rhabdias macrocephalum n. sp. Codon families (in alphabetical order, from left to right) are provided below the horizontal axis. Values at the top of each bar represent amino acid usage in percentage. |
The 36 gene arrangement in the mitogenomes of R. macrocephalum n. sp. differs from any of the arrangement types reported so far for Nematoda. The arrangement in R. macrocephalum n. sp. is in the following order: cox1, nad3, tRNA-Pro, cox2, tRNA-His, rrnL, nad5, tRNA-Ala, tRNA-Met, tRNA-Cys, tRNA-Val, tRNA-Asp, tRNA-Gly, nad6, nad4L, tRNA-Trp, tRNA-Glu, rrnS, tRNA-Ser2, tRNA-Asn, tRNA-Tyr, nad1, atp6, tRNA-Lys, tRNA-Leu2, tRNA-Ser1, nad2, tRNA-Ile, tRNA-Arg, tRNA-Gln, tRNA-Phe, cytb, tRNA-Leu1, cox3, tRNA-Thr, nad4 (Fig. 6).
 |
Figure 6 Linearized representation of the nematode mitochondrial gene arrangement of nematodes. The non-coding regions are not indicated. |
Molecular phylogeny of Rhabdiasidae
Phylogenetic results based on the ITS + 28S sequence data using ML and BI methods are almost identical (Fig. 7). The representatives of Rhabdiasidae were divided into four large monophyletic clades (Clade I, II, III, and IV). Clade I comprises species of Neoentomelas, Kurilonema, and Serpentirhabdias. Among them, Neoentomelas and Kurilonema have a closer relationship than Serpentirhabdias. Clade II includes representatives of Entomelas. Clade III contains species of Pneumonema, which showed a sister relationship with Clade IV, representing Rhabdias. In the genus Rhabdias, R. macrocephalum n. sp. formed a most basal lineage (Fig. 7).
 |
Figure 7 Maximum likelihood (ML) inference and Bayesian inference (BI) based on the ITS + 28S sequence data showing the phylogenetic relationships of representatives of Rhabdiasidae. Caenorhabditis elegans Dougherty (Rhabditida: Rhabditidae) was chosen as the out-group. Bootstrap values ≥70 and Bayesian posterior probabilities values ≥0.90 are shown in the phylogenetic trees. Bold indicates Rhabdias macrocephalum n. sp. |
Discussion
In the genus Rhabdias, a total of 21 species have been reported from lizards worldwide [5, 19, 37, 48]. Among them, only four species of Rhabdias were recorded from the lizards of the family Agamidae, including R. japalurae Kuzmin, 2003, R. singaporensis Bursey, Hoong & Goldberg, 2012, R. mcguirei Tkach, Kuzmin & Brown, 2011, and R. odilebaini Kuzmin, Tkach & Bush, 2012 [5, 19, 21, 48]. Rhabdias macrocephalum n. sp. can be easily distinguished from R. singaporensis by having a much longer esophagus (0.87–0.98 mm long, representing 5.35–6.00% of body length in R. macrocephalum vs 0.497–0.689 mm long, representing approximately 4.00% of body length in the latter) and different location of the excretory pore (just posterior to the nerve ring in the new species vs at the level of esophageal-intestinal junction in R. singaporensis) [5]. The new species is also different from R. odilebaini by having a particular pattern of cuticular inflation (cuticular inflation very narrow or inconspicuous in the anteriormost part and distinctly widening posteriorly from the level of nerve ring or mid-length of esophagus in the new species vs cuticle distinctly inflated to form a vesicle swollen in the anteriormost part of the body) and distinctly shorter tail (0.26–0.33 mm long, representing 1.52–2.02% of body length in the new species vs 0.36–0.50 mm long, representing 3.10–3.50% of body length in R. odilebaini) [19].
With the particular pattern of cuticular inflation, R. macrocephalum n. sp. is very similar to R. japalurae reported from Diploderma polygonatum Hallowell and D. swinhonis (Gunther) in Japan (Okinawa Island) and China (Taiwan Island), and R. mcguirei reported from Draco spilopterus (Wiegmann) in the Philippines [21, 48]. However, R. macrocephalum n. sp. can be differentiated from R. japalurae by having a distinctly shorter esophagus (0.87–0.98 mm long, representing 5.35–6.00% of body length in the new species vs 0.92–1.04 mm long, representing approximately 8.90–9.40% of body length in R. japalurae) [21]. The new species also differs from R. mcguirei by having a relatively shorter esophagus (esophageal length representing 5.35–6.00% of body length in the new species vs esophageal length representing 7.40–14.1% of body length in R. mcguirei) and different morphology of the tail (tail with distinct cuticular inflation and abruptly tapering from approximately 1/2 of region vs a tail with very narrow or inconspicuous cuticular inflation and abruptly tapering from anterior 1/3 of the region) [48]. Moravec [37] described R. lacerate Moravec, 2010 from the common lizard Lacerta vivipara Jacquin (Squamata: Lacertidae) in north-western Slovakia. This species with a very small body length (only 1.22–1.34 mm) and unique morphology of the tail tip (possessing 3 small cuticular spikes), is different from R. macrocephalum n. sp. Moreover, the other Rhabdias spp. reported from lizards are all collected from chameleonid and polychrotid hosts and distributed in tropical Africa, Madagascar, and Central America [3, 4, 19, 24–27, 32]. Additionally, R. macrocephalum n. sp. differs from all of these 21 Rhabdias spp. reported from lizards, including the four species parasitic in agamids, by having a conspicuously inflated cephalic extremity.
Molecular analyses of the partial 28S, ITS, cox1, and 12S sequences of R. macrocephalum n. sp. displayed no nucleotide divergence among different individuals, but showed a high level of genetic divergence between this new species and other Rhabdias spp. in these genetic makers, which also supports the hypothesis that the present material represents a new species of Rhabdias. Rhabdias macrocephalum n. sp. represents the ninth species of Rhabdias reported in China.
The current mitogenomic database for rhabdiasid nematodes remains very limited. Recently, the complete mitogenomes of R. kafunata and R. bufonis have been sequenced [52], which represented the only two rhabdiasid species with the mitogenomic data reported. The composition of the mitogenome of R. macrocephalum n. sp. [including 12 PCGs (missing atp8), 22 tRNA genes, and 2 rRNA genes] is identical to that of R. kafunata and R. bufonis, but the size of the complete mitogenome of R. macrocephalum n. sp. (14,819 bp) is slightly smaller than that of R. kafunata (15,437 bp) and R. bufonis (15,128 bp). Moreover, there are only three non-coding regions in the mitogenome of R. macrocephalum n. sp., but R. kafunata and R. bufonis have six and four non-coding regions in their mitogenomes, respectively. The mitogenomes of R. macrocephalum n. sp., R. kafunata, and R. bufonis all displayed a strong nucleotide compositional bias toward A + T (75.8–77.5%). To date, there have been 62 types of gene arrangements reported for the mitogenomes of nematodes [52]. The mitogenome of R. macrocephalum n. sp. showed a high level of gene rearrangement, which is different from that of R. kafunata, R. bufonis, and all of other mitogenomes of nematodes available so far, and represented a novel type of gene arrangement reported in Nematoda.
Recently, Zeng et al. [52] provided a basic molecular phylogenetic framework for the Rhabdiasidae based on ITS + 28S sequence data, and determined the systematic position of the Rhabdiasidae in the order Rhabditida using mitogenomic phylogeny. The present phylogenetic results agreed well with this study [52] and also supported the monophyly of Entomelas, Pneumonema, Serpentirhabdias, and Rhabdias. It is interesting that the present phylogenetic results displayed R. macrocephalum n. sp. forming a most basal lineage in the genus Rhabdias, being a sister to all other Rhabdias species. In the present phylogeny, only R. nicaraguensis Bursey, Goldberg & Vitt, 2007 was collected from a lizard host [4]; however, this species nested in these Rhabdias species collected from amphibians in South and North America, and did not display a close affinity with the new species. Additionally, R. macrocephalum n. sp. showed a distant relationship to the Eurasian Rhabdias species (i.e., R. bufonis, R. kafunata, R. nipponica, R. kongmonthaensis, R. bulbicauda, and R. bermani). The patterns of parasite–host switching and geographical distributions during the evolutionary history of Rhabdias ancestors is still an unsolved mystery. A more rigorous molecular phylogenetic study that includes broader representatives of Rhabdias species, especially these species collected from lizard hosts, is need to solve the above-mentioned issue.
Acknowledgments
The authors are grateful to the local veterinarian Mr. Huang for providing nematode specimens. This study was supported by the National Natural Science Foundation of China (Grant No. 32170442), the National Parasitic Resources Center and the Ministry of Science and Technology fund (NPRC-2019-194-30), and the National Key R&D Program of China (Grant No. 2022YFC2601200).
Conflicts of interest
The authors declare that they have no competing interests.
References
- Alcantara EP, Müller MI, Úngari LP, Ferreira-Silv C, Emmerich E, Giese EG, Morais DH, Santos ALQ, O’Dwyer LH, da Silva RJ. 2023. Integrative taxonomy in the genus Rhabdias Stiles & Hassall, 1905 from anuran in Brazil, description of two new species and phylogenetic analyses. Parasitology International, 93, e102714. [CrossRef] [PubMed] [Google Scholar]
- Bernt M, Merkle D, Ramsch K, Fritzsch G, Perseke M, Bernhard D, Schlegel M, Stadler PF, Middendorf M. 2007. Inferring genomic rearrangements based on common intervals. Bioinformatics, 23, 2957–2958. [CrossRef] [PubMed] [Google Scholar]
- Bursey CR, Goldberg SR, Telford SR. 2003. Rhabdias anolis n. sp. (Nematoda: Rhabdiasidae) from the lizard, Anolis frenatus (Sauria: Polychrotidae) from Panama. Journal of Parasitology, 89, 113–117. [CrossRef] [PubMed] [Google Scholar]
- Bursey CR, Goldberg SR, Vitt LJ. 2007. New species of Rhabdias (Nematoda: Rhabdiasidae) and other helminths from Norops capito (Sauria: Polychrotidae) from Nicaragua. Journal of Parasitology, 93, 129–131. [CrossRef] [PubMed] [Google Scholar]
- Bursey CR, Hoong DC, Goldberg SR. 2012. A new species of Rhabdias (Nematoda: Rhabdiasidae) in Calotes versicolor (Squamata: Agamidae) from Singapore. Journal of Parasitology, 98, 149–151. [CrossRef] [PubMed] [Google Scholar]
- Casiraghi M, Bain O, Guerrero R, Martin C, Pocacqua V, Gardner SL, Franceschi A, Bandi C. 2004. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: evidence for symbiont loss during evolution. International Journal for Parasitology, 34, 191–203. [CrossRef] [PubMed] [Google Scholar]
- Dare OK, Nadler SA, Forbes MR. 2008. Nematode lung-worms of two species of anuran amphibians: evidence for co-adaptation. International Journal for Parasitology, 38, 1729–1736. [CrossRef] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294–299. [PubMed] [Google Scholar]
- Gruber AR, Bernhart SH, Lorenz R. 2015. The viennarna web services. RNA Bioinformatics: Methods in Molecular Biology, 1269, 307–326. [CrossRef] [PubMed] [Google Scholar]
- Hasegawa H, Sato A, Kai M, Uchida A. 2013. Helminth parasites of bullfrogs Lithobates catesbeianus (Shaw, 1802) in Kanto District, Japan, with special reference to those introduced from North America. Japanese Journal of Veterinary Parasitology, 12, 1–10. [Google Scholar]
- Imai MD, Nadler AS, Brenner D, Donovan TA, Pessier A. 2009. Rhabditid nematode-associated ophthalmitis and meningoencephalomyelitis in captive Asian horned frogs (Megophrys montana). Journal of Veterinary Diagnostic Investigation, 21, 568–573. [CrossRef] [PubMed] [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology, 21, 241. [CrossRef] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [CrossRef] [Google Scholar]
- Kung CC, Wu HW. 1945. Parasitic nematodes of amphibians from Pehpei Szechwan, China. Sinensia Contributions from the Institute of Zoology Academia Sinica, 16, 73–83. [Google Scholar]
- Kuzmin Y, du Preez LH, Nel T, Svitin R. 2022. Three new species of Rhabdias Stiles & Hassall, 1905 (Nematoda: Rhabdiasidae) parasitic in Ptychadena spp. (Amphibia: Anura: Ptychadenidae) and an identification key to Rhabdias spp. from Afrotropical anurans. Parasitology International, 91, e102649. [CrossRef] [PubMed] [Google Scholar]
- Kuzmin Y, Halajian A, Tavakol S, Luus-Powell WJ, Tkach VV. 2017. Description and phylogenetic position of a new species of Rhabdias Stiles & Hassall, 1905 (Nematoda: Rhabdiasidae) from the banded rubber frog, Phrynomantis bifasciatus (Smith) (Amphibia: Microhylidae) in South Africa. Folia Parasitologica, 64, 35. [CrossRef] [Google Scholar]
- Kuzmin Y, Svitin R, Harnoster F, du Preez LH. 2020. Description and molecular characterisation of a new nematode species parasitic in the lungs of Strongylopus grayii (Smith) (Anura: Pyxicephalidae) in South Africa. Systematic Parasitology, 97, 369–378. [CrossRef] [PubMed] [Google Scholar]
- Kuzmin Y, Tkach VV, Brooks DR. 2007. Two new species of Rhabdias (Nematoda: Rhabdiasidae) from the marine toad, Bufo marinus (L.) (Lissamphibia: Anura: Bufonidae) in Central America. Journal of Parasitology, 93, 159–165. [CrossRef] [PubMed] [Google Scholar]
- Kuzmin Y, Tkach VV, Bush SE. 2012. A new species of Rhabdias (Nematoda: Rhabdiasidae) from agamid lizards on Luzon Island, Philippines. Journal of Parasitology, 98, 608–611. [CrossRef] [PubMed] [Google Scholar]
- Kuzmin Y, Tkach VV, Melo FTV. 2019. Description, molecular characterization and life cycle of Serpentirhabdias mussuranae n. sp. (Nematoda: Rhabdiasidae) from Clelia clelia (Reptilia: Colubroidea) in Brazil. Journal of Helminthology, 94, e55. [Google Scholar]
- Kuzmin Y. 2003. Rhabdias japalurae sp. nov. (Nematoda, Rhabdiasidae) from the japalures (Reptilia, Agamidae) and some notes on other Rhabdias spp. from lizards. Acta Parasitologica, 48, 6–11. [Google Scholar]
- Kuzmin Y. 2013. Review of Rhabdiasidae (Nematoda) from the Holarctic. Zootaxa, 3639, 1–76. [CrossRef] [PubMed] [Google Scholar]
- Lee BD. 2018. Python implementation of codon adaptation index. Journal of Open Source Software, 3, 905. [CrossRef] [Google Scholar]
- Lhermitte-Vallarino N, Bain O. 2004. Morphological and biological study of Rhabdias spp. (Nematoda) from African chameleons with description of a new species. Parasite, 11, 15–31. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Lhermitte-Vallarino N, Barbuto M, Ineich I, Wanji S, Lebreton M, Chirio L, Bain O. 2008. First report of Rhabdias (Nematoda: Rhabdiasoidea) from lungs of montane chameleons in Cameroon: description of two new species and notes on biology. Parasite, 15, 553–564. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Lhermitte-Vallarino N, Barbuto M, Junker K, Boistel R, Bain O. 2010. Rhabdias (Nematoda: Rhabdiasidae) from Chamaeleonidae (Sauria): two new species from Trioceros ellioti in East Africa and one from Brookesia superciliaris in Madagascar. Parasite, 17, 91–105. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Lhermitte-Vallarino N, Junker K, Bain O. 2009. Reappraisal of the specific status of Rhabdias (Nematoda: Rhabdiasoidea) from Malagasy chameleons in the Paris museum collection. Parasite, 16, 111–123. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Li YX, Huang XH, Li MR, Li SY, Huang ZJ, Wang DF, Yin GW, Wang L. 2023. Characterization and phylogenetic analysis of the complete mitochondrial genome of Rhabdias kafunata (Rhabditida: Rhabdiasidae). Experimental Parasitology, 255, 108646. [CrossRef] [Google Scholar]
- Lu SC. 1934. On Rhabdias, a genus of parasitic nematoda of Nanking. Sinensia, 5, 164–172. [Google Scholar]
- Machado SA, Kuzmin YI, Tkach VV, dos Santos JN, Gonçalves EC, de Vasconcelos Melo FT. 2018. Description, biology and molecular characterisation of Serpentirhabdias moi n. sp. (Nematoda: Rhabdiasidae) from Chironius exoletus (Serpentes: Colubridae) in Brazil. Parasitology International, 67, 829–837. [CrossRef] [PubMed] [Google Scholar]
- Marcaida AJB, Nakao M, Fukutani K, Nishikawa K, Urabe M. 2022. Phylogeography of Rhabdias spp. (Nematoda: Rhabdiasidae) collected from Bufo species in Honshu, Shikoku, and Kyushu, Japan including possible cryptic species. Parasitology International, 90, e102612. [CrossRef] [PubMed] [Google Scholar]
- Martínez-Salazar EA. 2006. A new Rhabdiasid species from Norops megapholidotus (Sauria: Polychrotidae) from Mexico. Journal of Parasitology, 92, 1325–1329. [CrossRef] [PubMed] [Google Scholar]
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Research, 47, e63. [CrossRef] [PubMed] [Google Scholar]
- Minh BQ, Hahn MW, Lanfear R. 2020. New methods to calculate concordance factors for phylogenomic datasets. Molecular Biology and Evolution, 37, 2727–2733. [CrossRef] [PubMed] [Google Scholar]
- Morais DH, Aguiar A, Müller MI, Narciso RB, da Silva LAF, da Silva RJ. 2016. Morphometric and phylogenetic analyses of Serpentirhabdias viperidicus n. sp. (Nematoda: Rhabdiasidae) from the lancehead snake Bothrops moojeni Hoge, 1966 (Reptilia: Serpentes: Viperidae) in Brazil. Journal of Helminthology, 91, 360–370. [Google Scholar]
- Morais DH, Müller MI, Melo FTV, Aguiar A, Willkens Y, de Sousa Silva C, Giese EG, Ávila RW, da Silva RJ. 2020. A new species of Rhabdias (Nematoda: Rhabdiasidae), a lung parasite of Pseudopaludicola pocoto (Anura: Leptodactylidae) from north-eastern Brazil: description and phylogenetic analyses. Journal of Helminthology, 94, e209. [CrossRef] [PubMed] [Google Scholar]
- Moravec F. 2010. Rhabdias lacertae n. sp. (Nematoda: Rhabdiasidae), the first rhabdiasid species parasitising lizards in Europe. Systematic Parasitology, 77, 23–27. [CrossRef] [PubMed] [Google Scholar]
- Müller MI, Morais DH, Costa-Silva GJ, Aguiar A, Ávila RW, da Silva RJ. 2018. Diversity in the genus Rhabdias (Nematoda, Rhabdiasidae): Evidence for cryptic speciation. Zoologica Scripta, 47, 595–607. [CrossRef] [Google Scholar]
- Müller MI, Morais DH, da Costa LFST, de Vasconcelos Melo FT, Giese EG, Ávila RW, da Silva RJ. 2023. Revisiting the taxonomy of Rhabdias fuelleborni Travassos, 1928 (Nematoda, Rhabdiasidae) with approaches to delimitation of species and notes on molecular phylogeny. Parasitology International, 92, e102692. [CrossRef] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, e129. [CrossRef] [Google Scholar]
- Ronquist F, Teslenko M, Mark PVD, Ayres DL, Darling A, Hhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. [CrossRef] [PubMed] [Google Scholar]
- Sata N, Nakano T. 2022. Insights into the phylogenetic position and phylogeography of the monospecific skink-parasite genus Neoentomelas (Nematoda: Rhabditida: Rhabdiasidae), with special reference to the effects of the reproductive mode on the genetic diversity. Invertebrate Systematics, 36, 36–47. [CrossRef] [Google Scholar]
- Sata N, Takeuchi H, Nakano T. 2020. A new species of Rhabdias (Nematoda: Rhabditida: Rhabdiasidae) from Miyakojima Island, Okinawa, Japan. Species Diversity, 25, 117–121. [CrossRef] [Google Scholar]
- Sonnenberg R, Nolte AW, Tautz D. 2007. An evaluation of LSU rDNA D1–D2 sequences for their use in species identification. Frontiers in Zoology, 4, e6. [CrossRef] [Google Scholar]
- Svitin R, Kuzmin YI, Preez LD. 2018. Molecular and morphological characterisation of Rhabdias picardiae Junker, Lhermitte-Vallarino et Bain, 2010 (Nematoda: Rhabdiasidae) from Delaland’s River frog, Amietia delalandii (Duméril et Bibron, 1841) (Amphibia: Pyxicephalidae) in South Africa. Acta Parasitologica, 63, 55–64. [CrossRef] [PubMed] [Google Scholar]
- Tavares-Costa LFS, Rebêlo GL, Müller MI, Jesus RF, Nandyara B, Silva LM, Costa-Campos CE, dos Santos JN, Melo FT. 2022. A new species of Rhabdias (Nematoda: Rhabdiasidae), a lung parasite of Pristimantis chiastonotus (Anura: Strabomantidae) from the Brazilian Amazon: description and phylogenetic analyses. Parasitology Research, 121, 155–166. [CrossRef] [PubMed] [Google Scholar]
- Tkach VV, Halajian A, Kuzmin Y. 2014. Phylogenetic affinities and systematic position of Entomelas sylvestris Baker, 1982 (Nematoda: Rhabdiasidae), a parasite of Breviceps sylvestris Fitz-Simons (Amphibia: Brevicipitidae) in South Africa. Systematic Parasitology, 87, 293–298. [CrossRef] [PubMed] [Google Scholar]
- Tkach VV, Kuzmin YI, Brown RM. 2011. Rhabdias mcguirei sp. nov. (Nematoda, Rhabdiasidae) from the flying lizard, Draco spilopterus (Squamata, Agamidae) of the northern Philippines. Acta Parasitologica, 56, 406–411. [Google Scholar]
- Tkach VV, Kuzmin YI, Pulis EE. 2006. A new species of Rhabdias from lungs of the wood frog, Rana sylvatica, in North America: the last sibling of Rhabdias ranae? Journal of Parasitology, 92, 631–636. [CrossRef] [PubMed] [Google Scholar]
- Tkach VV, Kuzmin YI, Snyder SD. 2014. Molecular insight into systematics, host associations, life cycles and geographic distribution of the nematode family Rhabdiasidae. International Journal for Parasitology, 44, 273–284. [CrossRef] [PubMed] [Google Scholar]
- Wang PQ, Wang YY, Zhao YR, Yan RL, Wang SP. 1992. Parasitic worms of vertebrates in and around the Meihua Mountain Nature Reserve. Wuyi Science Journal, 9, 31–48. (In Chinese). [Google Scholar]
- Zeng JL, Chen HX, Ni XF, Kang JY, Li L. 2024. Molecular phylogeny of the family Rhabdiasidae (Nematoda: Rhabditida), with morphology, genetic characterization and mitochondrial genomes of Rhabdias kafunata and R. bufonis. Parasites & Vectors, 17, e100. [CrossRef] [Google Scholar]
Cite this article as: Zeng J-L, Chen H-X, Xu H-R & Li L. 2024. Morphology, complete mitochondrial genome, and molecular phylogeny of Rhabdias macrocephalum n. sp. (Nematoda: Rhabdiasidae) from Diploderma splendidum (Reptilia: Agamidae). Parasite 31, 48.
All Tables
The primers and cycling conditions for amplifying different target regions by polymerase chain reaction (PCR) in the present study.
Detailed information on the representatives of Rhabdiasidae with their genetic data included in the phylogenetic analyses.
Annotations and gene organization of Rhabdias macrocephalum n. sp. Positive number in the “Gap or overlap” column indicates the length of intergenic sequence, and the negative number indicates the length (absolute number) that adjacent genes overlap (negative sign). The forward strand is marked as “+” and the reverse strand as “−”.
All Figures
 |
Figure 1 Photomicrographs of Rhabdias macrocephalum n. sp. from Diploderma splendidum in China. A: entire body (vulva arrowed), lateral view; B: anterior part of body, lateral view; C: region of vulva, lateral view; D: cephalic extremity, lateral view; E: posterior part of body (cloaca arrowed), lateral view. |
| In the text | |
 |
Figure 2 Line drawings of Rhabdias macrocephalum n. sp. from Diploderma splendidum in China. A: anterior part of body, lateral view; B: posterior part of body, lateral view; C: cephalic extremity, lateral view; D: cephalic extremity, apical view; E: region of vulva, lateral view; F: eggs. |
| In the text | |
 |
Figure 3 Scanning electron micrographs of Rhabdias macrocephalum n. sp. from Diploderma splendidum in China. A: anterior part of body, lateral view; B: tail, ventral view; C: magnified image of lateral cuticular pore; D: cephalic extremity (single papilla on each lip arrowed), apical view; E: magnified image of amphid; F: mid-body at level of vulva, sublateral view; G: egg with developed larva; H: magnified image of lateral cuticular pores on the tail; I: magnified image of lateral cuticular pores on the middle of body. Abbreviations: sl, submedian lip; ll, lateral lip. |
| In the text | |
 |
Figure 4 Gene maps of the mitochondrial genomes of Rhabdias macrocephalum n. sp. Abbreviations: NCR, non-coding region; PCG, protein coding gene; rRNA, ribosomal RNA; tRNA, transfer RNA. |
| In the text | |
 |
Figure 5 Relative synonymous codon usage (RSCU) of Rhabdias macrocephalum n. sp. Codon families (in alphabetical order, from left to right) are provided below the horizontal axis. Values at the top of each bar represent amino acid usage in percentage. |
| In the text | |
 |
Figure 6 Linearized representation of the nematode mitochondrial gene arrangement of nematodes. The non-coding regions are not indicated. |
| In the text | |
 |
Figure 7 Maximum likelihood (ML) inference and Bayesian inference (BI) based on the ITS + 28S sequence data showing the phylogenetic relationships of representatives of Rhabdiasidae. Caenorhabditis elegans Dougherty (Rhabditida: Rhabditidae) was chosen as the out-group. Bootstrap values ≥70 and Bayesian posterior probabilities values ≥0.90 are shown in the phylogenetic trees. Bold indicates Rhabdias macrocephalum n. sp. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


