| Issue |
Parasite
Volume 32, 2025
|
|
|---|---|---|
| Article Number | 37 | |
| Number of page(s) | 14 | |
| DOI | https://doi.org/10.1051/parasite/2025030 | |
| Published online | 24 June 2025 | |
Supplementary material
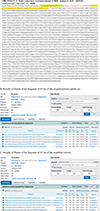 |
File S1. A. The original version of the primers-probe set of Homo sapiens ribonuclease P (RP forward: AGATTTGGACCTGCGAGCG; RP probe: TTCTGACCTGAAGGCTCTGCGCG; RP reverse: GAGCGGCTGTCTCCACAAGT) designed by Fan et al. [11] is shown in red letters and the modified version (RP-human forward: TCAGCATGGCGGTGTTT; RP-human probe: TTCTGACCTGAAGGCTCTGCGC; RP-human reverse: CGGCTGTCTCCACAAGTC) is in yellow highlight. Both sets were designed from the reference sequence (GenBank accession number): NM_006413.4. B. Results of Blastn of the fragment of 65 bp of the original primers-probe set. C. Results of Blastn of the fragment of 81 bp of the modified version. |
 |
File S2. Representative of optimization for the multiplex qPCR assay using primers-probe concentration for each target at (i) 250 nM primers/100 nM probe, (ii) 125 nM primers/50 nM probe, and (iii) 62.5 nM primers/25 nM probe and 102 fg/reaction for (A) L. martiniquensis, (B) L. orientalis, and (C) Crithidia sp. (CLA-KP1) as templates. The qPCR reactions were conducted according to the recommended thermal cycling protocol: 1 cycle of polymerase activation at 95 °C for 5 min, followed by 45 cycles of PCR at 95 °C for 15 s, and 60 °C for 1 min. |
|
File S3. PCR products of the ITS1 fragment of 149 bp using ITS1-L. mar primers, the ITS1 fragment of 146 bp using ITS1-L. ori/cha primers, and the ITS1 conserved fragment of 77 bp using ITS1-Tryps primers. Lanes: MW = Molecular weight markers; 1 = Negative control; 2 = human DNA; 3 = L. martiniquensis; 4 = L. orientalis; 5 = L. chancei; 6 = L. infantum; 7 = L. braziliensis; 8 = Crithidia sp. (CLA-KP1); 9 = L. martiniquensis + human DNA; 10 = L. orientalis + human DNA. |
|
File S4. Receiver operating characteristic (ROC) analysis for cut-off estimation for (A) L. martiniquensis, (B) L. orientalis, and (C) Crithidia sp. (CLA-KP1). |
|
File S5. Limit of detection (LOD95) of the multiplex qPCR for (A) L. martiniquensis, (B) L. orientalis, and (C) Crithidia sp. (CLA-KP1) was estimated by fitting a logistic regression model with detection results. Each dot indicates an actual detection rate at each dilution. The dashed line in the figure indicates the dilution ratio corresponding to a 95% probability. |
Table S1. Selected sequences of ITS1 targets of trypanosomatids that were used to design the primers and probes in this study. Access here
Table S2. Clinical diagnosis in the 69 residual DNA samples: 44 positive (42 for L. martiniquensis and 2 for L. orientalis) samples extracted from clinical specimens of leishmaniasis patients and 25 negative samples using the multiplex qPCR assay. Access here
© C. Mano et al., published by EDP Sciences, 2025
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


