| Issue |
Parasite
Volume 30, 2023
|
|
|---|---|---|
| Article Number | 62 | |
| Number of page(s) | 14 | |
| DOI | https://doi.org/10.1051/parasite/2023064 | |
| Published online | 19 December 2023 | |
Research Article
Prevalence of Spiroplasma and interaction with wild Glossina tachinoides microbiota
Prévalence de Spiroplasma et interaction avec le microbiote des Glossina tachinoides sauvages
1
Insect Pest Control Laboratory, Joint FAO/IAEA Centre of Nuclear Techniques in Food and Agriculture, 1400 Vienna, Austria
2
Insectarium de Bobo Dioulasso – Campagne d’Eradication de la mouche tsetse et de la Trypanosomose (IBD-CETT), 01 BP 1087, Bobo Dioulasso 01, Burkina Faso
3
Université Gaston Berger, Saint Louis, Senegal
4
Epidemiology, Parasites and Vectors, Agricultural Research Council-Onderstepoort Veterinary Research (ARC-OVR), Pretoria, South Africa
5
University of Dedougou, B.P. 176, Dédougou 01, Burkina Faso
6
Laboratory of Parasitology and Ecology, Faculty of Sciences, University of Yaounde I, Po. Box 812, Yaoundé, Cameroon
7
Centre for Research in Infectious Diseases (CRID), Po. Box 13591, Yaoundé, Cameroon
8
Institute of Chemical, Environmental, and Bioscience Engineering, Research Area Biochemical Technology, Vienna University of Technology, Gumpendorfer Straße 1a, 1060 Vienna, Austria
* Corresponding author: A.M.M.Abd-Alla@iaea.org
Received:
4
September
2023
Accepted:
28
November
2023
Tsetse flies (Diptera: Glossinidae) are vectors of the tropical neglected diseases sleeping sickness in humans and nagana in animals. The elimination of these diseases is linked to control of the vector. The sterile insect technique (SIT) is an environment-friendly method that has been shown to be effective when applied in an area-wide integrated pest management approach. However, as irradiated males conserve their vectorial competence, there is the potential risk of trypanosome transmission with their release in the field. Analyzing the interaction between the tsetse fly and its microbiota, and between different microbiota and the trypanosome, might provide important information to enhance the fly’s resistance to trypanosome infection. This study on the prevalence of Spiroplasma in wild populations of seven tsetse species from East, West, Central and Southern Africa showed that Spiroplasma is present only in Glossina fuscipes fuscipes and Glossina tachinoides. In G. tachinoides, a significant deviation from independence in co-infection with Spiroplasma and Trypanosoma spp. was observed. Moreover, Spiroplasma infections seem to significantly reduce the density of the trypanosomes, suggesting that Spiroplasma might enhance tsetse fly’s refractoriness to the trypanosome infections. This finding might be useful to reduce risks associated with the release of sterile males during SIT implementation in trypanosome endemic areas.
Résumé
Les mouches tsé-tsé (Diptera : Glossinidae) sont les vecteurs de maladies tropicales négligées, la maladie du sommeil chez l’homme et la nagana chez les animaux. L’élimination de ces maladies est liée à la lutte contre le vecteur. La technique de l’insecte stérile (TIS) est une méthode respectueuse de l’environnement qui s’est révélée efficace lorsqu’elle est appliquée dans le cadre d’une approche de lutte antiparasitaire intégrée à l’échelle d’une zone. Cependant, comme les mâles irradiés conservent leur compétence vectorielle, il existe un risque potentiel de transmission des trypanosomes lors de la libération des mâles sur le terrain. L’analyse de l’interaction entre la mouche tsé-tsé et son microbiote, et entre différents microbiotes et le trypanosome, pourrait fournir des informations importantes pour améliorer la résistance de la mouche à l’infection trypanosomienne. Cette étude sur la prévalence de Spiroplasma dans les populations sauvages de sept espèces de glossines d’Afrique de l’Est, de l’Ouest, centrale et australe a montré que Spiroplasma est présent uniquement chez Glossina fuscipes fuscipes et Glossina tachinoides. Chez G. tachinoides, un écart significatif par rapport à l’indépendance dans la co-infection par Spiroplasma et Trypanosoma spp. a été observé. De plus, les infections à Spiroplasma semblent réduire considérablement la densité des trypanosomes, ce qui suggère que Spiroplasma pourrait renforcer le caractère réfractaire de la mouche tsé-tsé aux infections trypanosomiennes. Cette découverte pourrait être utile pour réduire le risque associé à la libération de mâles stériles lors de la mise en œuvre de la TIS dans les zones d’endémie trypanosomienne.
Key words: Trypanosoma spp. / Microbe infection rate / Interactions / Paratransgenesis
© K.-S.M. Dera et al., published by EDP Sciences, 2023
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Tsetse flies (Diptera: Glossinidae) transmit trypanosomes, the causative agent of one of the most neglected vector-borne diseases in sub-Saharan Africa, i.e., African animal trypanosomosis or AAT (also called nagana) and human African trypanosomosis or HAT (sleeping sickness) [9, 37]. Tsetse flies are principally hematophagous and feed exclusively on vertebrate blood [2, 57]. During a blood meal on an infected host, the fly can ingest the trypanosomes which are established in the midgut. After several series of proliferation and differentiation, the trypanosomes mature in the salivary gland or the mouth parts depending on the trypanosome species. The parasite can then be transmitted to a mammalian host during a subsequent blood meal [60, 61].
The lack of effective prophylactic drugs or a vaccine [9], and the development of resistance to trypanocidal drugs [17], makes tsetse control the most efficient alternative for sustainable management of these diseases. One effective method for tsetse control is the Sterile Insect Technique (SIT) that needs to be implemented as part of an area-wide integrated pest management (AW-IPM) approach. The sterile insect technique requires mass production of the target insect, sterilization with irradiation and the release of these sterile insects in the field to mate with wild females to reduce fertility of the targeted population. However, the irradiation does not affect the tsetse fly’s susceptibility to develop mature trypanosome infections [18], and hence the desirability to enhance refractoriness of tsetse flies for trypanosome infections that would be used for release in an SIT program [59].
Symbiotic associations have been described in insects and typically involve bacteria that are vertically transmitted through progeny and may influence several functions of their hosts [51]. Tsetse flies harbor tree major endosymbiotic bacteria, i.e., the obligate mutualist Wigglesworthia glossinidia, the mutualist Sodalis glossinidius, and the parasitic Wolbachia pipientis [42]. Recently, a fourth endosymbiont, i.e., Spiroplasma was discovered in some natural tsetse populations and laboratory colonies of Glossina palpalis palpalis, Glossina fuscipes fuscipes, and Glossina tachinoides, all three being members of the palpalis group [21, 28]. In addition, multilocus sequencing typing (MLST) analysis identified two different strains of Spiroplasma in G. f. fuscipes and G. tachinoides [21].
Bacteria belonging to the genus Spiroplasma are Gram-positive, wall-less and are described in arthropods and plants. They are classified into three major monophyletic groups based on the 16S ribosomal RNA gene (rDNA) sequence: Ixodetis, Citri-Chrysopicola-Mirum (CCM), and Apis [24, 27]. They belong to the class of Mollicutes and are characterized by an helical shape and the lack of a cell wall and are only enveloped by a cholesterol-containing cell membrane [26]. The Spiroplasma are unique in having a well-defined, dynamic, helical cell geometry and a flat, monolayered, membrane-bound cytoskeleton, which follows, intracellularly, the shortest helical line on the cellular coil. They have a cytoskeleton which controls both the dynamic helical shape and the consequent motility of the cell [47, 58]. Their cell size varies between 100 and 240 nm [1]. The genome size ranges from 780 to 2,220 kb and is rich in AT [1, 47]. The role of Spiroplasma in the tsetse fly host is currently unclear, but it has been reported to have a negative effect on the viability of Harmonia axyridis, and male killing activity in Drosophila melanogaster and Drosophila neotestacea [5, 24]. Many studies have also revealed that Spiroplasma might cause disease in arthropods and plants [5, 39]. Conversely, some Spiroplasma strains might have a positive effect in their hosts conferring resistance against pathogens [34, 41, 46]. Like Wolbachia, Spiroplasma can be found in ovaries. However, they primarily reside in the hemolymph but can also be detected in fat body and salivary glands. Using a laboratory colony of G. f. fuscipes, it was shown that Spiroplasma may interact with trypanosomes [50]. Flies harboring Spiroplasma presented a lower prevalence of trypanosome infection in the midgut, indicating a potential negative correlation between Spiroplasma presence and trypanosome infection. In the same study, it was shown that Spiroplasma can be transmitted vertically, although the possibility of horizontal transmission could not be excluded. These findings supported the use of Spiroplasma in paratransgenesis approaches to develop trypanosome refractoriness. The use of paratransgenesis has been suggested as an approach that could confer resistance against pathogens by genetically engineering the symbionts or the vector [7, 14]. This approach has been implemented successfully in triatome bugs [22] and mosquitoes [62], but is still under evaluation for tsetse flies. In this respect, the use of the endosymbiont Sodalis was previously recommended for paratransgenic approaches in tsetse flies [6, 15, 16]. In support of this idea, irradiating 22 day-old pupae did not impact the copy number of Sodalis in G. morsitans morsitans compared to non-irradiated flies [18]. However, it has been reported that Sodalis has a negative impact on the metabolic and reproductive fitness of G. f. fuscipes [56].
In this study, the prevalence of Spiroplasma infection was assessed in natural tsetse populations collected from different countries in Africa. The potential interaction between Spiroplasma with the trypanosomes and Wigglesworthia was studied using a G. tachinoides population from Burkina Faso and Ghana. We also report on the genotyping and presence of different Spiroplasma strains in wild G. tachinoides populations.
Materials and methods
Tsetse taxon collection and DNA purification
Wild populations of tsetse flies were collected in 40 locations in 10 different countries in West Africa (Burkina Faso, Ghana, Guinea, Mali and Senegal), Central Africa (Democratic Republic of Congo), East Africa (Ethiopia and Uganda) and southern Africa (South Africa and Zimbabwe). Eight tsetse taxa were analysed, including G. brevipalpis, G. f. fuscipes, G. m. morsitans, Glossina morsitans submorsitans, Glossina pallidipes, Glossina palpalis gambiensis, Glossina palpalis palpalis and G. tachinoides (Fig. 1, Supplementary Table 1).
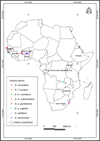 |
Figure 1 Geographical locations of tsetse samples in Africa. |
Adult flies were collected in 1995 and between 2005 and 2018 using the biconical Challier-Laveissière trap, the monoconical Vavoua trap [11, 13], the Ngu trap, the Epsilon trap baited with acetone [25], and the odour baited H trap [32]. The collected flies were stored in 95% absolute ethanol or propylene glycol and shipped to the Insect Pest Control Laboratory (IPCL) of the Joint FAO/IAEA Centre of Nuclear Techniques in Food and Agriculture, Seibersdorf, Austria. At the IPCL, the samples were stored at −20 °C until further use. Total DNA was extracted from the whole body of each individual fly using a DNeasy tissue kit (QIAGEN Inc., Valencia, CA, USA), following the supplier’s instructions.
Prevalence of Spiroplasma and Trypanosoma
To detect Spiroplasma infection, PCR amplification of an approximately 455 bp fragment of the 16S rRNA gene was performed [21]. The PCR was carried out in 25 μL reaction mixtures containing 22.5 μL of 1.1× Pre-Aliquoted PCR Master Mix (0.625 units Thermoprime Plus DNA Polymerase, 75 mM Tris–HCl (pH 8.8 at 25 °C), 20 mM (NH4)2SO4, 2.0 mM MgCl2, 0.01% (v/v) Tween-20 and 0.2 mM each of the dNTPs (ABgene, UK), and 1.5 μL of template DNA plus 1 μL of Spiroplasma 16S RNA primers (63F and TKSS) (Supplementary Table 2) to a final concentration of 0.2 mM per primer. PCR conditions were 95 °C for 5 min, followed by 34 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, and final extension 72 °C for 10 min. PCR products were electrophoresed in 2% molecular grade agarose (Fisher Biotech) stained with SafeGreen. DNA from the IPCL colony of G. f. fuscipes, which is known to be infected with Spiroplasma, and sterilized distilled water were included in each PCR test as positive and negative controls, respectively. As described previously [19], the Glossina species microsatellite GpCAG133 was used to control the quality of the extracted DNA and only validated samples were considered for Spiroplasma or Trypanosoma infection status. To confirm that the amplified PCR products obtained with G. pallidipes, G. m. morsitans, and G. p. gambiensis were Spiroplasma-specific sequences, two approaches were used: the first was to confirm the specificity of the amplification by performing PCR using the MLST primers shown in Supplementary Table 2. The second was to sequence the PCR product obtained by the 16S rRNA gene primers. For the sequencing, PCR products were purified using the High Pure PCR Clean-up Micro Kit (Roche, Basel, Switzerland) and ligated to the pGEM-T vector (Promega, Madison, WI, USA), following the supplier’s instructions. The recombinant plasmids were transformed into DH5α-competent bacteria (Invitrogen, Carlsbad, CA, USA), following the supplier’s instructions. The recombinant plasmids and the inserted sequences were confirmed by Sanger sequencing (Eurofins Genomics, Ebersberg, Germany) with the universal vector primers M13F_uni (-21) (5′–TGT AAA ACG GCC AGT–3′) and M13R_rev (-29) (5′–CAG GAA ACA GCT ATG ACC–3′). For the other tsetse species including G. f. fuscipes, G. brevipalpis and G. tachinoides, the amplified PCR products were purified with the ZR-96 DNA Clean & Concentrator®-5 (Zymo Research, Irvine, CA, USA), following the manufacturer’s protocol and submitted directly to sequencing without cloning using the 63F and TKSS primers (Eurofins Genomics, Ebersberg, Germany). The resulting sequences were blasted against the non-redundant protein sequence (nr) database in the NCBI server using the BLAST tool https://blast.ncbi.nlm.nih.gov/Blast.cgi to identify and annotate the sequence. The sequence was considered a Spiroplasma sequence if it matched with Spiroplasma sequence in the database. The prevalence of Trypanosoma was assessed as in Ouedraogo et al. [43].
Analysis of the interaction between Spiroplasma and Trypanosoma in G. tachinoides
Co-infection of Spiroplasma and Trypanosoma spp. was evaluated by PCR. Trypanosome prevalence was determined as previously described [43]. Infection status was divided into four categories: 1. Spiroplasma positive and Trypanosoma positive (Sp+/T+), 2. Spiroplasma positive and Trypanosoma negative (Sp+/T−), 3. Spiroplasma negative and Trypanosoma positive (Sp−/T+), and 4. Spiroplasma negative and Trypanosoma negative (Sp−/T−).
Analysis of the density of Spiroplasma, Trypanosoma and Wigglesworthia infection density
Samples showing the following infection status (Sp+/T+), (Sp+/T−), and (Sp+/T−) were used to assess the density of Spiroplasma, Trypanosoma spp., and Wigglesworthia using relative quantitative PCR (qPCR). The qPCR was performed using a CFX96 Real Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The Spiroplasma density was assessed by the amplification of 16S rRNA gene with the qPCR Spiroplasma primers (Supplementary Table 2). In addition, the density of Wigglesworthia was evaluated as previously described [18] using the thiC (thiamine biosynthesis gene) (Supplementary Table 2). Based on the above-mentioned criteria, 212 G. tachinoides individuals (76, 65 and 71 flies with (Sp+/T+), (Sp+/T−), and (Sp−/T+) infection status, respectively) were selected from Burkina Faso and Ghana samples. In addition, samples with (Sp+/Tryp+), (Sp+/T−), and (Sp−/T+) were used to assess the impact of Spiroplasma infection on Trypanosoma density. Trypanosomatidae 18S rRNA gene specific primers (18S_Typ_F and 18S_Typ_R) (Supplementary Table 2) were used to assess the Trypanosoma density in the tested samples. The DNA from all selected samples was diluted to a final concentration of 4 ng/μL and 5 μL of the diluted DNA was used for qPCR to determine Spiroplasma, Wigglesworthia, and Trypanosoma DNA density normalized to the housekeeping β-tubulin gene. The amplification mixture contained 5 μL of DNA template, 200 nM of each primer, and 7.5 μL iQTM SYBER Green Supermix (Bio-Rad). qPCR cycling conditions for Spiroplasma and Wigglesworthia were as follows: initial denaturation at 95 °C for 2 min; 39 cycles of 95 °C for 5 s, 55 °C for 30 s, one step at 95 °C for 5 s, and a melting curve constructed from 65 °C to 95 °C in increments of 0.5 °C for 5 s. The same conditions were used for Trypanosoma, except the annealing temperature, which was 60 °C. Analysis of the Spiroplasma, Wigglesworthia, Trypanosoma, and β-tubulin densities was based only on the qPCR data with the expected melting curve at 81.5 °C, 85.5 °C, and 86 °C, respectively.
Genetic variation and phylogenetic analysis of Spiroplasma in G. tachinoides
To assess the genetic variation of Spiroplasma in wild G. tachinoides, an MLST approach was employed on positive samples from Burkina Faso and Ghana using the following genes: 16S rRNA, Spiroplasma fructose repressor (fruR), Spiroplasma DNA Topoisomerase 4 subunit B (parE), and RNA polymerase subunit beta (rpoB). Primer sets used for each reaction, product sizes, and PCR conditions are shown in Supplementary Table 2.
All amplified PCR products were purified using a High Pure PCR Cleanup Micro Kit (Roche Diagnostics, Indianapolis, IN, USA) and a ZR-96 DNA Clean-up Kit™ (Zymo Research, Irvine, CA, USA). Sequencing was performed with Eurofins Genomics Company (https://www.eurofinsgenomics.com) and sequencing data were first analyzed using Geneious Prime® 2023.0.2 and then blasted using the “Blast” resource of NCBI to confirm them as Spiroplasma. Phylogenetic trees were built for each gene (16S rRNA, fruR, pare, and rpoB) and for the concatenated data set using all four gene sequences. Multiple alignments were then performed using MUSCLE alignment with the default parameters on Geneious Prime® 2023.0.2 and the Neighbor-joining tree was built using the Tamura-Nei genetic distance model.
Data analysis
Prevalence data for Spiroplasma and Trypanosoma spp. were analysed in R using Rstudio version V 1.4.1106 [4] and general linear model (glm) [45] combined with analysis of variance, respectively provided by the ggplot package [63] to detect potential differences between countries and between localities in each country. Spiroplasma, Trypanosoma, and Wigglesworthia density levels were first normalized with the tsetse house-keeping β-tubulin gene and only the samples for which density levels were available for all three microorganisms were used for further analysis. The analysis was performed with the glm to detect the significant differences between the density of Spiroplasma, Trypanosoma, and Wigglesworthia according to the Spiroplasma and Trypanosoma co-infection status (Supplementary File 1). To evaluate the association between Spiroplasma and Trypanosoma, the Cochran-Manthel-Haenzel (CMH) test and the chi-square test were performed on the excel table, as described previously [19].
Results
Prevalence of Spiroplasma
The presence of Spiroplasma in wild populations of tsetse flies was assessed using a PCR-based method to amplify part of the 16S rRNA gene. Positive samples were identified based on the observed amplicon band size in the electrophoresis gel for all tsetse species. Sequencing of the respective PCR amplicons revealed that Spiroplasma infection was only confirmed in G. tachinoides (N = 41) and G. f. fuscipes (N = 6), both belonging to the palpalis subgenus (Table 1). In the case of G. brevipalpis, G. m. morsitans, G. m. submorsitans, G. pallidipes, G. p. gambiensis, and G. p. palpalis, the amplified sequence belonged to different microbial species, primarily Bacillus cereus, Bacillus thuringiensis, Enterococcus cecorum, and some uncultured bacteria (Data not shown).
Spiroplasma identification in 8 Glossina species from 10 different countries using 16S rRNA, multilocus sequence typing (MLST), and Sanger sequencing.
The PCR results indicated an overall Spiroplasma prevalence of 39.27% in G. tachinoides. The prevalence did not differ significantly between Burkina Faso, Ghana, and the laboratory colony (χ2 = 2.12, df = 2, and p = 0.34), with Burkina Faso and Ghana showing a prevalence rate of 46.56% and 52.94%, respectively (Table 2). However, a significant variation in Spiroplasma prevalence was found across the various sampling locations (χ2 = 22.61, df = 8, and p = 0.003) (Table 2 and Figs. 2 and 3). Specifically, there was a significant difference in prevalence between the two sampling locations in Burkina Faso (χ2 = 6.459, df = 1, and p = 0.01), with a higher prevalence observed in Folonzo. Similarly, a significant difference was found between the prevalence rate in different locations in Ghana (χ2 = 11.955, df = 5, and p = 0.03), with the highest prevalence observed in the Mortani region (98.44%), where 100% of the female flies were infected. Conversely, the lowest prevalence of Spiroplasma was recorded in Kumpole, Ghana (25%), with male flies showing no sign of infection (Table 2, Figs. 2 and 3).
 |
Figure 2 Prevalence of Spiroplasma according to location. Bars marked with the same lower-case letter do not differ significantly at the 0.05 level. |
 |
Figure 3 Prevalence of Spiroplasma and Trypanosoma (single and multiple) infections per country, location, and sex. Prevalence data were square root transformed and averaged based on location-sex and the matrix display was conducted in PRIMER version 7 + software. Tree on the left of the matrix is the similarity dendrogram based on the similarity index of the square root of the prevalence values. The color index is the square root of the prevalence values ranged 0–9 which is the square root of 0–81% prevalence. |
Global prevalence of Spiroplasma in Glossina tachinoides according to locations and countries.
Prevalence of single and multiple Trypanosoma infections
The screening of the flies indicated the presence of different taxa of Trypanosoma, including Tc (Trypanosoma congolense type: Savanah, Kilifi, Forest), Tv (Trypanosoma vivax), and Tz (Trypanozoon sp.: Trypanosoma brucei brucei, Trypanosoma brucei gambiense, Trypanosoma brucei rhodesiense, Trypanosoma evansi). The overall prevalence of single or multiple Trypanosoma infections among all tested flies was 69.97% (457/653). The prevalence of Trypanosoma varied significantly between countries (χ2 = 37.18, df = 1, and p < 0.001) and locations (χ2 = 452.21, df = 7, and p < 0.001). In Ghana, the prevalence was significantly higher than in Burkina Faso, at 86.38% and 20.76%, respectively (Table 2 and Fig. 3). In Ghana, the prevalence varied significantly with location (χ2 = 125.43, df = 5, and p < 0.001), with a prevalence of 100% in some locations such as Sissili Bridge, Fumbissi, Kumpole, and Grogro (Fig. 3 and Supplementary Table 3).
The most frequently found trypanosomes were Tz and Tv, with a prevalence of 30.2% and 22.42%, respectively. However, only Tz varied significantly with country (χ2 = 7.54, df = 1, and p = 0.006) and location (χ2 = 185.82, df = 7, and p < 0.001). Trypanosoma congolense was found in the two locations in Burkina Faso (Comoe at 2.37% and Folonzo at 2.00%), and only in one location in Ghana (Walewale (2.87%)). Its prevalence varied significantly with country (χ2 = 6.426, df = 1, and p = 0.01) and location (χ2 = 34.97, df = 7, and p < 0.001).
The TvTz multiple infection was the most prevalent in the samples (11.22%). In Ghana, no TcTv double infections were found, while in Burkina Faso, no triple infections TcTvTz were found. The prevalence of the double infections varied only according with location (χ2 = 245.15, df = 7, and p < 0.001) (Fig. 3 and TcTz Supplementary Table 3).
Interaction between Spiroplasma and Trypanosoma
Prevalence of co-infections
The results of the analysis showed that 12.56% of the flies were infected both with Spiroplasma and Trypanosoma, regardless of country, location, and sex. However, the prevalence of single infections of Spiroplasma (35.83%) was higher than that of Trypanosoma (17.46%) (Fig. 4). The association between Spiroplasma and Trypanosoma infections was analyzed using the Cochran-Manthel-Haenzel (CMH) test and chi-square test. Across all samples, the CMH test showed a significant deviation from independence between the two infections (χ2MH = 5.19, df = 1, p = 0.02). The chi-square test confirmed that the independence between Spiroplasma and Trypanosoma infections was significant with a Bonferroni correction of α = 0.006 (χ2 = 9.85, p = 0.03). However, when considering countries, only in Ghana the chi-square test did show a significant deviation from independence between the two microbial infections (χ2 = 13.004, p < 0.001) (Table 3 and Supplementary Table 4).
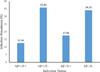 |
Figure 4 Prevalence of co-infection Spiroplasma-Trypanosoma in wild G. tachinoides. |
Distribution of the association between the presence of Trypanosoma spp. and the presence of Spiroplasma according to country and location.
Co-infection and the density of Spiroplasma, Trypanosoma, and Wigglesworthia
The density of Spiroplasma, Trypanosoma, and Wigglesworthia was evaluated using relative qPCR based on the single (Sp+/T−; Sp−/T+) and double co-infection (Sp+/T+) status. As expected, the results showed that flies infected with Spiroplasma (Sp+/T− and Sp+/T+) had a significantly higher density of Spiroplasma compared to those not infected (Sp−/T+), which indicated that flies classified as uninfected by conventional PCR showed lower infection rates with qPCR. However, there was no significant difference in the density of Spiroplasma between flies infected with Spiroplasma and not infected with Trypanosoma (Sp+/T−) and those infected with both (Sp+/T+) (Fig. 5A). Furthermore, flies with double co-infection (Sp+/T+) had a significantly higher density of trypanosomes than those with single co-infection (Sp+/T− and Sp−/T+) (Fig. 5B). However, no significant difference was found in the density of Wigglesworthia in the three categories of co-infection (Fig. 5C).
 |
Figure 5 Normalized density of Spiroplasma (A), Trypanosoma (B), and Wigglesworthia (C) according to Spiroplasma-Trypanosoma co-infection in wild G. tachinoides. Bars marked with the same lower-case letter do not differ significantly at the 0.05 level. |
Genetic variation and phylogenetic analysis of Spiroplasma in wild G. tachinoides
Among the 35 samples sequenced, 14 sequences from Comoe in Burkina Faso, two from the CIRDES colony, and two from Walewale in Ghana were used for the analysis. For the four genes used for the sequencing, 2,885 base pairs of sequence were generated. The comparison of the sequences showed a global nucleotide mutation rate of 0.06% with two SNPs (Table 4). These two SNPs were found on the parE gene (1SNP/745 bp) and rpoB gene (1SNP/1455). None of these substitutions were non-synonymous and the percentage of amino-acid mutations was 0.40% (1/248) for the parE gene and 0.20% (1/485) for the rpoB gene. For the parE gene, the mutation resulted in the replacement of isoleucine to valine, but for the rpoB gene from phenylalanine to serine. All samples from all locations showed the same profile for the 16S rRNA and fruR genes. In Burkina Faso and Ghana, two genotypes were found, while only one was detected for CIRDES (Tables 5 and 6). Three different haplotypes were found in the sampling areas with a specific haplotype for the CIRDES colony and Burkina Faso and Ghana sharing the same haplotypes (Table 6, Fig. 6).
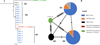 |
Figure 6 Neighbor-Joining consensus tree (A) and Haplotype network analysis (B) of the Spiroplasma in G. tachinoides in Burkina Faso and Ghana. (A) Neighbor-Joining consensus tree was built after alignment of all the concatenated sequences. The method used to calculate the distance was Tamura-Nei. (B) Haplotype network generated based on the ML tree which was generated based on Spiroplasma sequences. The black lineaments on the lines represent mutation events between the haplotypes. The different colors represent the locations. The reference sequence of Spiroplasma in G. fuscipes fuscipes species (KX159391) was used as the outgroup for construction of both phylogenetic tree and haplotype network. |
Summary of information on nucleotide polymorphisms detected in the partial sequences of Spiroplasma in G. tachinoides.
Alleles of Spiroplasma in different locations of tested countries. Numbers between brackets indicate the number of sequences tested per each allele.
Spiroplasma haplotypes found in the same individuals collected in East and Southern African countries. The frequency of occurrence of the haplotypes is shown in the last column. The number in parentheses indicates the total number of flies in which the haplotype was detected.
Discussion
In this study, we evaluated the prevalence of the endosymbiont Spiroplasma and the Trypanosoma parasite in wild G. tachinoides in Burkina Faso and Ghana, and the interaction between these two microbes and Wigglesworthia. The discovery of the presence of Spiroplasma in tsetse flies is quite recent, although its presence in other insects and plants has been known for long time. Doudoumis et al. [21] showed the presence of Spiroplasma in both laboratory colonies and field populations of G. tachinoides, G f. fuscipes, and G. p palpalis, all belonging to the palpalis group.
The present study confirmed the presence of Spiroplasma in G. tachinoides in both wild populations and colonized insectary flies. Using 16S rRNA gene sequencing, we observed amplification of a bacterial community, different from Spiroplasma, in the tsetse species that did not belong to the palpalis group. Since the 16S rRNA gene is shared with all bacterial species and is one of the most conserved bacterial genes [64], primers designed to target this region could have detected a broad range of bacteria species. It is therefore necessary to carry out taxonomical confirmation by sequencing the respective amplicons [30]. The prevalence in the field was found to be similar to that observed by El Khamlichi et al. [23], but higher than that reported by Doudoumis et al. [21]. However, the prevalence of Spiroplasma in the colony was lower than that observed by Doudoumis et al. [21]. The prevalence of the infection varied significantly with location. Furthermore, although the prevalence rates did not differ significantly between Burkina Faso and Ghana, the observed differences in prevalence rates between individual sampling locations suggest that regional variations may impact the infection and the distribution of Spiroplasma.
In the study area, three major trypanosomes of humans and animals were found, T. brucei s.l. (Tz), T. congolense (Tc), and T. vivax (Tv), with a relatively high prevalence, particularly in Ghana. This high prevalence of Trypanosoma in the flies explains the presence of AAT in the sampling site, highlighting the significant risk of infection in this area. A human infection risk cannot be excluded as Tz was identified. Indeed, they are the main cause of HAT [10, 37]. The presence of Tv in Burkina Faso was already shown previously [55]. The prevalence of Trypanosoma was almost similar to the result of Djohan et al. [20] in Côte d’Ivoire (61.4%), but significantly higher (69.97%) than the prevalence obtained by Kame-Ngasse et al. [31] in the north of Cameroon (34.81%) and Meharenet and Alemu [38] in Ethiopia. The difference in prevalence compared to the results obtained by Meharenet and Alemu [38] could be due to the diagnostic method used. In their study, the authors used dissection to identify the presence or absence of the parasite, which has some disadvantages, including low sensitivity and susceptibility to the examiner’s technical expertise [12]. Female flies appeared to have higher infection rates than males, which is in line with the results of Meharenet and Alemu [38] and Lefrançois et al. [33], and may be due to their longer lifespan. Trypanozoon sp and T. vivax were the most predominant Trypanosoma. Djohan et al. [20] also found the same predominant species of trypanosome with Tv present at 27.2%. Conversely, Kame-Ngasse et al. [31] observed that in G. tachinoides in Cameroon, Tc was dominant. Since the active foci of the HAT are different, the distribution of parasites will depend on the sampling area. Previous studies have shown the predominance of Tc in the “Faro and Deo” region in Cameroon [35, 36].
Mixed infections were predominantly TvTz (11.22%) and TcTz (4.45%). Previous studies have shown that G. tachinoides is commonly infected with various types of trypanosomes. However, the composition of the mixed infections may depend on the distribution of the parasite and the identification method used. For instance, in Côte d’Ivoire, TvTcs (9.4%) and TcTcs (12.5%) were the predominant mixed infections [20], whereas in Cameroon TcsTcf (4.8%) was the predominant one [31].
Our analyses with relative qPCR showed that a Spiroplasma-Trypanosoma coinfection had no significant effect on the density of Wigglesworthia. This bacterium is an obligate tsetse fly endosymbiont that provides essential nutrients that are absent in blood meals [8]. It is maternally transmitted, making it difficult for other microbiota to invade its niche [3]. Although Spiroplasma can be maternally transmitted [56], its presence or absence did not affect the density of Wigglesworthia or Sodalis in laboratory G. f. fuscipes flies.
The analysis of the prevalence of Spiroplasma and Trypanosoma coinfections suggests a significant deviation from independence, as most of the flies infected with Spiroplasma were not infected with Trypanosoma, and vice versa. This may indicate that the presence of Spiroplasma could confer a certain level of refractoriness to Trypanosoma infection. This hypothesis was confirmed by the Cochran-Mantel-Haenszel (CMH) and chi-square tests, that showed a significant deviation from independence between the two microorganisms across all samples. Our results align with those of Schneider et al. [50] who reported that only 2% of Spiroplasma infected flies in G. f. fuscipes species were also infected with trypanosomes. The same study also found that, under laboratory conditions, trypanosomes were less likely to colonize the midgut of G. f. fuscipes infected with Spiroplasma. However, our results did not agree with the higher prevalence of Spiroplasma found in Trypanosoma-infected Glossina palpalis palpalis flies than uninfected ones [40]. The mechanism by which this bacterium enhances refractoriness to trypanosomes in flies remains unclear. It could be related to competition for proliferation niches, given that Spiroplasma is found in both the midgut and hemolymph, or to the induction of an immune response in the fly or specific gene regulation. It might also be due to competition for specific nutrients that both microbes need for their development. This has been observed with the endosymbiont Sodalis, which competes with the host and parasite for host nutrients [52–54]. Our study indicates that the possible refractory effect of Spiroplasma on trypanosome infection is not species-dependent, as it was observed in both G. tachinoides and G. f. fuscipes. However, these two species belong to the palpalis subgroup, within which Spiroplasma was exclusively found in our study. Our genotyping showed that the strains of Spiroplasma found in G. tachinoides in Burkina Faso and Ghana are most closely related to the citri group, as previously reported. This clade is composed of various taxa that are pathogens for plants, such as S. phoeniceum [49] and S. citri [48], as well as protecting Drosophila against nematode infection and parasitic wasps such as S. poulsonii [29, 44, 65, 66]. Despite belonging to the same citri group, three haplotypes were identified, with Burkina Faso and Ghana sharing two haplotypes and one specific haplotype for the CIRDES colony samples. This information sheds light on the genetic diversity of Spiroplasma in the field and in laboratory colonies, which could help us to understand its evolution. The colonization process may induce several mutations that could lead to the development of new haplotypes. Insect microbiota can be influenced by a variety of factors, including environmental conditions, host genetics, and interactions with other organisms. When an insect is colonized, its microbiota may be exposed to different environmental conditions or may interact with new microbial communities, resulting in changes in the composition or function of the microbiota.
The SIT for tsetse flies relies on the release of sterile males within the context of area-wide insect pest management (AW-IPM). To prevent or reduce the transmission of trypanosomes by the released sterile males, they receive at least two blood meals with trypanocidal drugs before being released. However, this is cumbersome and costly, so the discovery that Spiroplasma infection could confer refractoriness to the trypanosome infection in flies presents an elegant way to mitigate the transmission risk. Releasing sterile males infected with Spiroplasma, that are to a certain degree refractory to the trypanosome parasite, would reduce the risk of transmission. Moreover, since paternal transmission of Spiroplasma occurs, albeit imperfectly, the offspring from the residual fertility of the sterile males released could also be infected with Spiroplasma and be relatively refractory to the parasite. Spiroplasma is an endosymbiont that can significantly improve the effectiveness of SIT, making the study and management of this microbe crucial.
Conclusion
This study reinforced the hypothesis that Spiroplasma could enhance refractoriness to trypanosome infections in certain species of tsetse flies, and this would make this symbiont a good candidate for paratransgenesis in addition to Sodalis, as previously described. More investigations are required with field samples to better understand interactions between Spiroplasma and the trypanosome, and to evaluate the impact of ionizing radiation on the dynamism of Spiroplasma.
Funding
This study was supported by the Joint FAO/IAEA Insect Pest Control Subprogram and the IAEA’s Department of Technical Cooperation.
Data Availability Statement
Materials described in the paper, including all relevant raw data, are available at this link https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/ZKIR8I.
Acknowledgments
The authors would like to acknowledge colleagues and technicians at the insect pest control laboratory in Seibersdorf, Austria for providing orientation in the laboratory and supplying materials when needed. The authors thank the many collaborators in Africa who helped to collect tsetse fly samples.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary materials
Supplementary File 1: R-Marckdown file with details of the data analysis.
Supplementary Table 1: Details of the geographic coordinates of the sampling sites in Africa.
Supplementary Table 2: List of Primers used for PCR and quantitative PCR (qPCR) analyses of microbiome in Glossina tachinoides.
Supplementary Table 3: Prevalence in percentage of Spiroplasma, Trypanosoma spp., and the different Trypanosoma species, single or multiple infection in Burkina Faso and Ghana, according to sampling location and sex. Spiro = Spiroplasma, T. spp = Trypanosoma spp., Tc = T. congolense, Tv = T. vivax, Tz = Trypanosoma brucei spp., TcTv = Coinfection T. congolense - T. vivax, TcTz = Coinfection T. congolense - T. brucei spp., TvTz = Coinfection T. vivax - T. brucei spp., TcTvTZ = Coinfection T. congolense, T. vivax, and T. brucei spp. Prevalence in percentage of Spiroplasma, Trypanosoma spp., and the different Trypanosoma species, single or multiple infection in Burkina Faso and Ghana, according to sampling location and sex. Spiro = Spiroplasma, T. spp. = Trypanosoma spp., Tc = T. congolense, Tv = T. vivax, Tz = Trypanosoma brucei spp., TcTv = Coinfection T. congolense - T. vivax, TcTz = Coinfection T. congolense - T. brucei spp., TvTz = Coinfection T. vivax - T. brucei spp., TcTvTZ = Coinfection T. congolense, T. vivax, and T. brucei spp.
Supplementary Table 4: Chi-2 test of independence between Spiroplasma and Trypanosoma.
Access hereReferences
- Agrios GN. 2005. Chapter twelve – Plant diseases caused by prokaryotes: bacteria and mollicules, in Plant pathology, 5th edn. George N Agrios, Editor. Academic Press: San Diego. p. 615–703. [CrossRef] [Google Scholar]
- Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nature Genetics, 32, 402–407. [CrossRef] [PubMed] [Google Scholar]
- Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S. 2008. Analysis of milk gland structure and function in Glossina morsitans: Milk protein production, symbiont populations and fecundity. Journal of Insect Physiology, 54, 1236–1242. [CrossRef] [PubMed] [Google Scholar]
- Baier T, Neuwirth E. 2007. Excel :: COM :: R. Computational Statistics, 22, 91–108. [CrossRef] [Google Scholar]
- Bastian FO, Elzer PH, Wu X. 2012. Spiroplasma spp. biofilm formation is instrumental for their role in the pathogenesis of plant, insect and animal diseases. Experimental and Molecular Pathology, 93, 116–128. [CrossRef] [PubMed] [Google Scholar]
- Beard CB, O’Neill SL, Mason P, Mandelco L, Woese CR, Tesh RB, Richards FF, Aksoy S. 1993. Genetic transformation and phylogeny of bacterial symbionts from tsetse. Insect Molecular Biology, 1, 123–131. [CrossRef] [PubMed] [Google Scholar]
- Beaty BJ. 2000. Genetic manipulation of vectors: A potential novel approach for control of vector-borne diseases. Proceedings of the National Academy of Sciences of the United States of America, 97, 10295–10297. [CrossRef] [PubMed] [Google Scholar]
- Bing X, Attardo GM, Vigneron A, Aksoy E, Scolari F, Malacrida A, Weiss BL, Aksoy S. 2017. Unravelling the relationship between the tsetse fly and its obligate symbiont Wigglesworthia: transcriptomic and metabolomic landscapes reveal highly integrated physiological networks., 284, 20170360. [Google Scholar]
- Brun R, Blum J, Chappuis F, Burri C. 2010. Human African trypanosomiasis. Lancet, 375, 148–159. [CrossRef] [PubMed] [Google Scholar]
- Cecchi G, Mattioli RC, Slingenbergh J, de la Rocque S. 2008. Land cover and tsetse fly distributions in sub-Saharan Africa. Medical and Veterinary Entomology, 22, 364–373. [CrossRef] [PubMed] [Google Scholar]
- Challier A, Laveissière C. 1973. Un nouveau piège pour la capture des glossines (Glossina: Diptera, Muscidae): description et essais sur le terrain. Cahiers ORSTOM. Série Entomologie Médicale et Parasitologie, 11, 251–262. [Google Scholar]
- Cirad. 2004. The Educational Tsetse Fly La mouche tsé-tsé pédagogique. Les Savoirs partagés CIRAD: Montpellier, France. [Google Scholar]
- Claude L, Jean-Pierre E, Pascal G, Jean-Jacques L. 1990. The control of riverine tsetse. International Journal of Tropical Insect Science, 11, 427–441. [CrossRef] [Google Scholar]
- Coutinho-Abreu IV, Zhu KY, Ramalho-Ortigao M. 2010. Transgenesis and paratransgenesis to control insect-borne diseases: Current status and future challenges. Parasitology International, 59, 1–8. [CrossRef] [PubMed] [Google Scholar]
- De Vooght L, Caljon G, De Ridder K, van den Abbeele J. 2014. Delivery of a functional anti-trypanosome Nanobody in different tsetse fly tissues via a bacterial symbiont, Sodalis glossinidius. Microbial Cell Factories, 13, 156. [CrossRef] [PubMed] [Google Scholar]
- De Vooght L, Caljon G, Stijlemans B, de Beatselier P, Coosemans M, Van Den Abbeele J. 2012. Expression and extracellular release of a functional anti-trypanosome Nanobody (R) in Sodalis glossinidius, a bacterial symbiont of the tsetse fly. Microbial Cell Factories, 11, 23. [CrossRef] [PubMed] [Google Scholar]
- Delespaux V, Geysen D, Van den Bossche P, Geerts S. 2008. Molecular tools for the rapid detection of drug resistance in animal trypanosomes. Trends in Parasitology, 24, 236–242. [CrossRef] [PubMed] [Google Scholar]
- Demirbas-Uzel G, De Vooght L, Parker AG, Vreysen MJB, Mach RL, Van Den Abbeele J, Abd-Alla AMM. 2018. Combining paratransgenesis with SIT: impact of ionizing radiation on the DNA copy number of Sodalis glossinidius in tsetse flies. BMC Microbiology, 18, 160. [CrossRef] [PubMed] [Google Scholar]
- Dieng MM, Dera KM, Moyaba P, Ouedraogo GMS, Demirbas-Uzel G, Gstöttenmayer F, Mulandane FC, Neves L, Mdluli S, Rayaisse J-B, Belem AMG, Pagabeleguem S, de Beer CJ, Parker AG, Van Den Abbeele J, Mach RL, Vreysen MJB, Abd-Alla AMM. 2022. Prevalence of Trypanosoma and Sodalis in wild populations of tsetse flies and their impact on sterile insect technique programmes for tsetse eradication. Scientific Reports, 12, 3322. [CrossRef] [PubMed] [Google Scholar]
- Djohan V, Kaba D, Rayaissé J-B, Dayo G-K, Coulibaly B, Salou E, Dofini F, Kouadio ADMK, Menan H, Solano P. 2015. Detection and identification of pathogenic trypanosome species in tsetse flies along the Comoé River in Côte d’Ivoire. Parasite, 22, 18. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Doudoumis V, Blow F, Saridaki A, Augustinos AA, Dyer NA, Goodhead IB, Solano P, Rayaisse J-B, Takac P, Mekonnen S, Parker AG, Abd-Alla AMM, Darby AC, Bourtzis K, Tsiamis G. 2017. Challenging the Wigglesworthia, Sodalis, Wolbachia symbiosis dogma in tsetse flies: Spiroplasma is present in both laboratory and natural populations. Scientific Reports, 7, 4699. [CrossRef] [PubMed] [Google Scholar]
- Durvasula RV, Gumbs A, Panackal A, Kruglov O, Aksoy S, Merrifield RB, Richards FF, Beard CB. 1997. Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proceedings of the National Academy of Sciences of the United States of America, 94, 3274–3278. [CrossRef] [PubMed] [Google Scholar]
- El Khamlichi S, Maurady A, Asimakis E, Stathopoulou P, Sedqui A, Tsiamis G. 2022. Detection and characterization of Spiroplasma and Wolbachia in a natural population of Glossina tachinoides, in Adv. Intell. Syst. Sustain. Dev. AI2SD’2020. Kacprzyk J, Balas VE, Ezziyyani M, Editors. Springer International Publishing: Cham. p. 256–264. [Google Scholar]
- Goryacheva I, Blekhman A, Andrianov B, Romanov D, Zakharov I. 2018. Spiroplasma infection in Harmonia axyridis – Diversity and multiple infection. PLoS One, 13, e0198190. [CrossRef] [PubMed] [Google Scholar]
- Hargrove JW, Langley PA. 1990. Sterilizing tsetse (Diptera: Glossinidae) in the field: a successful trial. Bulletin of Entomological Research, 80, 397–403. [CrossRef] [Google Scholar]
- Harne S, Gayathri P, Béven L. 2020. Exploring Spiroplasma biology: Opportunities and challenges. Frontiers in Microbiology, 11, 589279. [CrossRef] [PubMed] [Google Scholar]
- Haselkorn TS, Markow TA, Moran NA. 2009. Multiple introductions of the Spiroplasma bacterial endosymbiont into Drosophila. Molecular Ecology, 18, 1294–1305. [CrossRef] [PubMed] [Google Scholar]
- Jacob F, Melachio TT, Njitchouang GR, Gimonneau G, Njiokou F, Abate L, Christen R, Reveillaud J, Geiger A. 2017. Intestinal bacterial communities of trypanosome-infected and uninfected Glossina palpalis palpalis from three human African trypanomiasis foci in Cameroon. Frontiers in Microbiology, 8, 1464. [CrossRef] [PubMed] [Google Scholar]
- Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science, 329, 212–215. [CrossRef] [PubMed] [Google Scholar]
- Jenkins C, Ling CL, Ciesielczuk HL, Lockwood J, Hopkins S, McHugh TD, Gillespie SH, Kibbler CC. 2012. Detection and identification of bacteria in clinical samples by 16S rRNA gene sequencing: comparison of two different approaches in clinical practice. Journal of Medical Microbiology, 61, 483–488. [CrossRef] [PubMed] [Google Scholar]
- Kame-Ngasse GI, Njiokou F, Melachio-Tanekou TT, Farikou O, Simo G, Geiger A. 2018. Prevalence of symbionts and trypanosome infections in tsetse flies of two villages of the “Faro and Déo” division of the Adamawa region of Cameroon. BMC Microbiology, 18, 159. [CrossRef] [PubMed] [Google Scholar]
- Kappmeier K. 2000. A newly developed odour-baited “H trap” for the live collection of Glossina brevipalpis and Glossina austeni (Diptera: Glossinidae) in South Africa. Onderstepoort Journal of Veterinary Research, 67, 15–26. [Google Scholar]
- Lefrançois T, Solano P, Bauer B, Kabore I, Touré SM, Cuny G, Duvallet G. 1999. Polymerase chain reaction characterization of trypanosomes in Glossina morsitans submorsitans and G. tachinoides collected on the game ranch of Nazinga, Burkina Faso. Acta Tropica, 72, 65–77. [CrossRef] [PubMed] [Google Scholar]
- Majerus TMO, Graf von der Schulenburg JH, Majerus MEN, Hurst GDD. 1999. Molecular identification of a male-killing agent in the ladybird Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae). Insect Molecular Biology, 8, 551–555. [CrossRef] [PubMed] [Google Scholar]
- Mamoudou A, Zoli A, Mbahin N, Tanenbe C, Bourdanne CP-H, Marcotty T, den Bossche PV, Geerts S. 2006. Prevalence and incidence of bovine trypanosomosis on the Adamaoua plateau in Cameroon 10 years after the tsetse eradication campaign. Veterinary Parasitology, 142, 16–22. [CrossRef] [PubMed] [Google Scholar]
- Mamoudou A, Zoli A, Tchoua P. 2010. Parasitological prevalence of bovine trypanosomosis in the Faro and Deo division valley of the Adamaoua plateau, Cameroon. International Journal of Biological and Chemical Sciences, 3, 1192–1197. [CrossRef] [Google Scholar]
- Mattioli RC, Feldmann U, Hendrickx G, Wint W, Jannin J, Slingenbergh J. 2004. Tsetse and trypanosomiasis intervention policies supporting sustainable animal-agricultural development. Journal of Food, Agriculture and Environment, 2, 310–314. [Google Scholar]
- Meharenet B, Alemu D. 2020. Trypanosome infection rate in Glossina tachinoides: infested rivers of Limmu Kosa District Jimma Zone. Western Ethiopia. BMC Research Notes, 13, 133. [CrossRef] [Google Scholar]
- Mouches C, Bové JM, Albisetti J, Clark TB, Tully JG. 1982. A Spiroplasma of serogroup IV causes a May-disease-like disorder of honeybees in Southwestern France. Microbial Ecology, 8, 387–399. [CrossRef] [PubMed] [Google Scholar]
- Ngambia Freitas FS, Njiokou F, Tsagmo Ngoune JM, Sempere G, Berthier D, Geiger A. 2021. Modulation of trypanosome establishment in Glossina palpalis palpalis by its microbiome in the Campo sleeping sickness focus, Cameroon. Infection, Genetics and Evolution, 90, 104763. [CrossRef] [PubMed] [Google Scholar]
- Oliver KM, Smith AH, Russell JA. 2014. Defensive symbiosis in the real world – advancing ecological studies of heritable, protective bacteria in aphids and beyond. Functional Ecology, 28, 341–355. [CrossRef] [Google Scholar]
- O’Neill SL, Gooding RH, Aksoy S. 1993. Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Medical and Veterinary Entomology, 7, 377–383. [CrossRef] [PubMed] [Google Scholar]
- Ouedraogo GMS, Demirbas-Uzel G, Rayaisse J-B, Gimonneau G, Traore AC, Avgoustinos A, Parker AG, Sidibe I, Ouedraogo AG, Traore A, Bayala B, Vreysen MJB, Bourtzis K, Abd-Alla AmM. 2018. Prevalence of trypanosomes, salivary gland hypertrophy virus and Wolbachia in wild populations of tsetse flies from West Africa. BMC Microbiology, 18, 153. [CrossRef] [PubMed] [Google Scholar]
- Paredes JC, Herren JK, Schüpfer F, Marin R, Claverol S, Kuo C-H, Lemaitre B, Béven L. 2015. Genome sequence of the Drosophila melanogaster male-killing Spiroplasma strain MSRO endosymbiont. mBio, 6, e02437-14. [CrossRef] [PubMed] [Google Scholar]
- R Core Team. 2021. R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Ratzka C, Gross R, Feldhaar H. 2012. Endosymbiont tolerance and control within insect hosts. Insects, 3, 553–572. [CrossRef] [PubMed] [Google Scholar]
- Regassa LB. 2014. The Family Spiroplasmataceae, in The Prokaryotes. Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, Editors. Springer: Berlin, Heidelberg. p. 551–567. [CrossRef] [Google Scholar]
- Saglio P, Lhospital M, Lafleche D, Dupont G, Bové JM, Tully JG, Freundt EA. 1973. Spiroplasma citri gen. and sp. n.: a mycoplasma-like organism associated with “stubborn” disease of citrus. International Journal of Systematic and Evolutionary Microbiology, 23, 191–204. [Google Scholar]
- Saillard C, Vignault JC, Bové JM, Raie A, Tully JG, Williamson DL, Fos A, Garnier M, Gadeau A, Carle P. 1987. Spiroplasma phoeniceum sp. nov., a new plant-pathogenic species from Syria. International Journal of Systematic and Evolutionary Microbiology, 37, 106–115. [Google Scholar]
- Schneider DI, Saarman N, Onyango MG, Hyseni C, Opiro R, Echodu R, O’Neill M, Bloch D, Vigneron A, Johnson TJ, Dion K, Weiss BL, Opiyo E, Caccone A, Aksoy S. 2019. Spatio-temporal distribution of Spiroplasma infections in the tsetse fly (Glossina fuscipes fuscipes) in northern Uganda. PLOS Neglected Tropical Diseases, 13, e0007340. [CrossRef] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature, 407, 81–86. [CrossRef] [PubMed] [Google Scholar]
- Snyder AK, Deberry JW, Runyen-Janecky L, Rio RVM. 2010. Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proceedings of the Royal Society B: Biological Sciences, 277, 2389–2397. [CrossRef] [PubMed] [Google Scholar]
- Snyder AK, Rio RV. 2013. Interwoven biology of the tsetse holobiont. Journal of Bacteriology, 195, 4322–4330. [CrossRef] [PubMed] [Google Scholar]
- Snyder AK, Rio RVM. 2015. Wigglesworthia morsitans folate (Vitamin B9) biosynthesis contributes to tsetse host fitness. Applied and Environmental Microbiology, 81, 5375–5386. [CrossRef] [PubMed] [Google Scholar]
- Solano P, Reifenberg JM, Amsler-Delafosse S, Kabore I, Cuisance D, Duvallet G. 1996. Trypanosome characterization by polymerase chain reaction in Glossina palpalis gambiensis and G. tachinoides from Burkina Faso. Medical and Veterinary Entomology, 10, 354–358. [CrossRef] [PubMed] [Google Scholar]
- Son JH, Weiss BL, Schneider DI, Dera KM, Gstöttenmayer F, Opiro R, Echodu R, Saarman NP, Attardo GM, Onyango M, Abd-Alla AMM, Aksoy S. 2021. Infection with endosymbiotic Spiroplasma disrupts tsetse (Glossina fuscipes fuscipes) metabolic and reproductive homeostasis. PLoS Pathogens, 17, e1009539. [CrossRef] [PubMed] [Google Scholar]
- Tobe SS, Langley PA. 1978. Reproductive physiology of Glossina. Annual Review of Entomology, 23, 283–307. [CrossRef] [PubMed] [Google Scholar]
- Trachtenberg S, Gilad R. 2002. A bacterial linear motor: cellular and molecular organization of the contractile cytoskeleton of the helical bacterium Spiroplasma melliferum BC3: A bacterial linear motor. Molecular Microbiology, 41, 827–848. [Google Scholar]
- Van Den Abbeele J, Bourtzis K, Weiss B, Cordón-Rosales C, Miller W, Abd-Alla AMM, Parker AG. 2013. Enhancing tsetse fly refractoriness to trypanosome infection – a new IAEA coordinated research project. Journal of Invertebrate Pathology, 112, S142–S147. [CrossRef] [PubMed] [Google Scholar]
- Van Den Abbeele J, Claes Y, Van Bockstaele D, Le Ray D, Coosemans M. 1999. Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology, 118, 469–478. [CrossRef] [PubMed] [Google Scholar]
- Vickerman K, Tetley L, Hendry KAK, Turner CMR. 1988. Biology of African trypanosomes in the tsetse fly. Biology of the Cell, 64, 109–119. [CrossRef] [PubMed] [Google Scholar]
- Wang S, Ghosh AK, Bongio N, Stebbings KA, Lampe DJ, Jacobs-lorena M. 2012. Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proceedings of the National Academy of Sciences of the United States of America, 109, 12734–12739. [CrossRef] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag: New York. [Google Scholar]
- Woese CR. 1987. Bacterial evolution. Microbiological Reviews, 51, 221–271. [CrossRef] [PubMed] [Google Scholar]
- Xie J, Butler S, Sanchez G, Mateos M. 2014. Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity, 112, 399–408. [CrossRef] [PubMed] [Google Scholar]
- Xie J, Vilchez I, Mateos M. 2010. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS One, 5, e12149. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Dera K-SM, Dieng MM, Moyaba P, Ouedraogo GMS, Pagabeleguem S, Njokou F, Freitas FSN, de Beer CJ, Mach RL, Vreysen MJB & Abd-Alla AMM. 2023. Prevalence of Spiroplasma and interaction with wild Glossina tachinoides microbiota. Parasite 30, 62.
All Tables
Spiroplasma identification in 8 Glossina species from 10 different countries using 16S rRNA, multilocus sequence typing (MLST), and Sanger sequencing.
Global prevalence of Spiroplasma in Glossina tachinoides according to locations and countries.
Distribution of the association between the presence of Trypanosoma spp. and the presence of Spiroplasma according to country and location.
Summary of information on nucleotide polymorphisms detected in the partial sequences of Spiroplasma in G. tachinoides.
Alleles of Spiroplasma in different locations of tested countries. Numbers between brackets indicate the number of sequences tested per each allele.
Spiroplasma haplotypes found in the same individuals collected in East and Southern African countries. The frequency of occurrence of the haplotypes is shown in the last column. The number in parentheses indicates the total number of flies in which the haplotype was detected.
All Figures
 |
Figure 1 Geographical locations of tsetse samples in Africa. |
| In the text | |
 |
Figure 2 Prevalence of Spiroplasma according to location. Bars marked with the same lower-case letter do not differ significantly at the 0.05 level. |
| In the text | |
 |
Figure 3 Prevalence of Spiroplasma and Trypanosoma (single and multiple) infections per country, location, and sex. Prevalence data were square root transformed and averaged based on location-sex and the matrix display was conducted in PRIMER version 7 + software. Tree on the left of the matrix is the similarity dendrogram based on the similarity index of the square root of the prevalence values. The color index is the square root of the prevalence values ranged 0–9 which is the square root of 0–81% prevalence. |
| In the text | |
 |
Figure 4 Prevalence of co-infection Spiroplasma-Trypanosoma in wild G. tachinoides. |
| In the text | |
 |
Figure 5 Normalized density of Spiroplasma (A), Trypanosoma (B), and Wigglesworthia (C) according to Spiroplasma-Trypanosoma co-infection in wild G. tachinoides. Bars marked with the same lower-case letter do not differ significantly at the 0.05 level. |
| In the text | |
 |
Figure 6 Neighbor-Joining consensus tree (A) and Haplotype network analysis (B) of the Spiroplasma in G. tachinoides in Burkina Faso and Ghana. (A) Neighbor-Joining consensus tree was built after alignment of all the concatenated sequences. The method used to calculate the distance was Tamura-Nei. (B) Haplotype network generated based on the ML tree which was generated based on Spiroplasma sequences. The black lineaments on the lines represent mutation events between the haplotypes. The different colors represent the locations. The reference sequence of Spiroplasma in G. fuscipes fuscipes species (KX159391) was used as the outgroup for construction of both phylogenetic tree and haplotype network. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


