Figure 6
Download original image
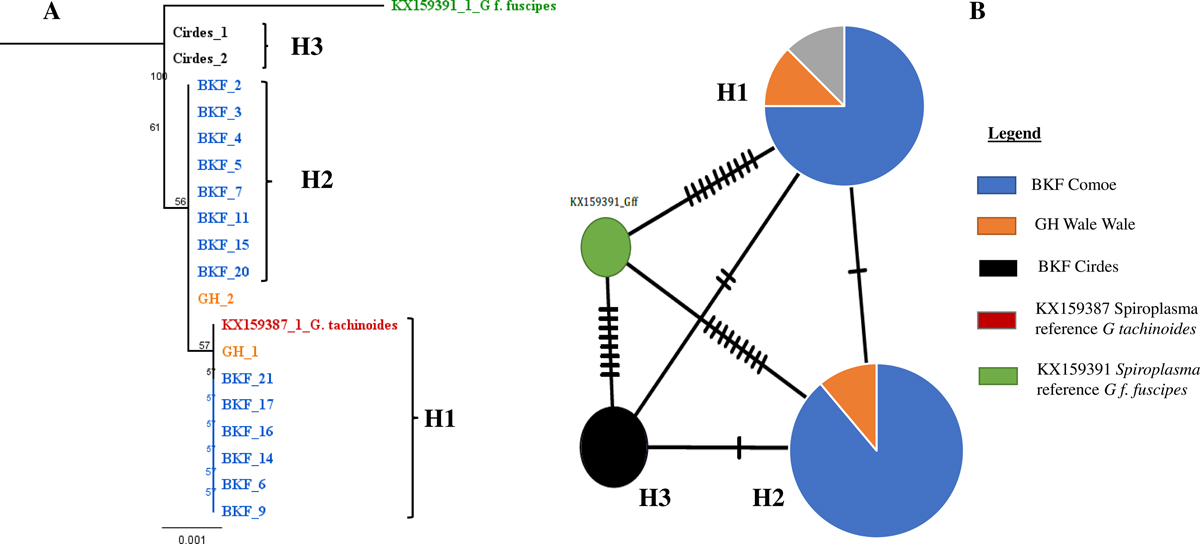
Neighbor-Joining consensus tree (A) and Haplotype network analysis (B) of the Spiroplasma in G. tachinoides in Burkina Faso and Ghana. (A) Neighbor-Joining consensus tree was built after alignment of all the concatenated sequences. The method used to calculate the distance was Tamura-Nei. (B) Haplotype network generated based on the ML tree which was generated based on Spiroplasma sequences. The black lineaments on the lines represent mutation events between the haplotypes. The different colors represent the locations. The reference sequence of Spiroplasma in G. fuscipes fuscipes species (KX159391) was used as the outgroup for construction of both phylogenetic tree and haplotype network.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


