| Issue |
Parasite
Volume 27, 2020
|
|
|---|---|---|
| Article Number | 46 | |
| Number of page(s) | 10 | |
| DOI | https://doi.org/10.1051/parasite/2020043 | |
| Published online | 20 July 2020 | |
Research Article
Molecular identification of trypanosomes in cattle in Malawi using PCR methods and nanopore sequencing: epidemiological implications for the control of human and animal trypanosomiases
Identification moléculaire des trypanosomes chez les bovins du Malawi, à l’aide de méthodes de PCR et du séquençage par nanopores : implication épidémiologique pour le contrôle des trypanosomiases humaines et animales
1
Division of Collaboration and Education, Research Center for Zoonosis Control, Hokkaido University, Kita-20, Nishi-10, Kita-ku, Sapporo, 001-0020 Hokkaido, Japan
2
Laboratory of Parasitology, Veterinary Medicine Faculty, Hokkaido University, Kita-18, Nishi-9, Kita-ku, Sapporo, 060-0818 Hokkaido, Japan
3
Department of Pathology, College of Medicine, University of Malawi, P/Bag 360 Chichiri, 30096 Blantyre 3, Malawi
4
International Collaboration Unit, Research Center for Zoonosis Control, Hokkaido University, Kita-20, Nishi-10, Kita-ku, Sapporo, 001-0020 Hokkaido, Japan
* Corresponding author: junya@czc.hokudai.ac.jp
Received:
4
December
2019
Accepted:
25
June
2020
This study aimed to identify trypanosomes infecting cattle in Malawi in order to understand the importance of cattle in the transmission dynamics of Human African Trypanosomiasis (HAT) and Animal African Trypanosomosis (AAT). A total of 446 DNA samples from cattle blood from three regions of Malawi were screened for African trypanosomes by ITS1 PCR. The obtained amplicons were sequenced using a portable next-generation sequencer, MinION, for validation. Comparison of the results from ITS1 PCR and MinION sequencing showed that combining the two methods provided more accurate species identification than ITS1 PCR alone. Further PCR screening targeting the serum resistance-associated (SRA) gene was conducted to detect Trypanosoma brucei rhodesiense. Trypanosoma congolense was the most prevalent Trypanosoma sp., which was found in Nkhotakota (10.8%; 20 of 185), followed by Kasungu (2.5%; 5 of 199). Of note, the prevalence of T. b. rhodesiense detected by SRA PCR was high in Kasungu and Nkhotakota showing 9.5% (19 of 199) and 2.7% (5 of 185), respectively. We report the presence of animal African trypanosomes and T. b. rhodesiense from cattle at the human–livestock–wildlife interface for the first time in Malawi. Our results confirmed that animal trypanosomes are important causes of anemia in cattle and that cattle are potential reservoirs for human African trypanosomiasis in Malawi.
Résumé
Cette étude visait à identifier les trypanosomes infectant les bovins au Malawi afin de comprendre l’importance des bovins dans la dynamique de transmission de la trypanosomiase humaine africaine (THA) et de la trypanosomose animale africaine (TAA). Au total, 446 échantillons d’ADN de sang de bovins provenant de trois régions du Malawi ont été soumis à un dépistage des trypanosomes africains par PCR de l’ITS1. Les amplicons obtenus ont été séquencés à l’aide d’un séquenceur portable de nouvelle génération, MinION, pour validation. La comparaison des résultats de la PCR de l’ITS1 et de la séquence MinION a montré que la combinaison des deux méthodes permettait une identification plus précise des espèces que la seule PCR de l’ITS1. Un autre dépistage par PCR ciblant le gène SRA (associé à la résistance du sérum) a été effectué pour détecter Trypanosoma brucei rhodesiense. Trypanosoma congolense était l’espèce de trypanosome la plus répandue, trouvée à Nkhotakota (10,8 % ; 20 sur 185), suivi de Kasungu (2,5 % ; 5 sur 199). Notamment, la prévalence de T. b. rhodesiense détectée par PCR de SRA était élevée à Kasungu et Nkhotakota, avec respectivement 9,5 % (19 sur 199) et 2,7 % (5 sur 185). Nous rapportons la présence de trypanosomes animaux africains et de T. b. rhodesiense de bovins à l’interface homme-bétail-faune sauvage, pour la première fois au Malawi. Nos résultats confirment que les trypanosomes animaux sont des causes importantes d’anémie chez les bovins et que les bovins sont des réservoirs potentiels pour la trypanosomiase humaine africaine au Malawi.
Key words: Cattle / Epidemiology / AAT / HAT / Malawi / Trypanosome
© M. Marsela et al., published by EDP Sciences, 2020
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
African trypanosomiasis is caused by protozoan parasites of Trypanosoma spp. and is mainly transmitted by tsetse flies [52]. This disease is a major concern in sub-Saharan Africa, with detrimental effects on both human and animal health and causing significant losses to affected countries [52]. Animal African trypanosomiasis (AAT) is caused by T. congolense, T. vivax, and T. brucei brucei [50]. Infection in domestic animals is usually severe, unlike in wildlife, where it is usually nonpathogenic [13]. AAT affects domestic animals, including cattle, goats, sheep, and pigs, and its pathogenicity differs according to the host species [13, 50]. Clinical symptoms include fever, anemia, loss of weight and productivity, abortion, decreased fertility, edema, paralysis, and even death [6]. AAT remains a major threat to animal health and stock farming within the tsetse belt [13]. Unlike T. congolense and T. brucei, which are transmitted by tsetse flies, T. vivax can also be transmitted mechanically by other hematophagous flies; as a result, it has a broader geographical distribution [44].
Human African trypanosomiasis (HAT) or sleeping sickness occurs in two forms with different features, due to T. b. gambiense and T. b. rhodesiense infections, respectively [8]. Gambiense HAT caused by T. b. gambiense is an anthroponotic disease that depends primarily on human-to-human transmission; humans act as the main reservoir, while animal reservoirs play a minor role [17]. It is distributed mostly in western and central Africa, an area that currently has 98% of reported cases of HAT [64]. Gambiense HAT is a chronic infection, during which a person can be infected for a long period of time without demonstrating major clinical signs of the disease. Symptoms often appear at the late stage of the disease, when the central nervous system is already affected [7]. In contrast, rhodesiense HAT caused by T. b. rhodesiense is a zoonotic disease that affects mainly animals (wildlife and livestock); humans are considered accidental hosts [17]. It is found in 13 countries in eastern and southern Africa, representing under 2% of reported cases of HAT, and it causes an acute infection [64]. The disease is known to progress quickly and invades the central nervous system right after onset of symptoms, a few months or weeks after infection [7].
Control of rhodesiense HAT is challenging because animals act as reservoirs for disease transmission [17, 63]. Although they do not show any clinical symptoms, animal reservoirs harbor parasites, and tsetse flies can acquire the infection [7]. Necessary control measures face obstacles because animal infections are difficult to monitor, unlike in humans, where they can be easily tracked. Wildlife have long been known to be the major reservoir of T. b. rhodesiense [41]. Livestock can also act as potent reservoir hosts for T. b. rhodesiense due to high exposure of humans to agriculture [61]. In Uganda, cattle were implicated as the principal domestic reservoirs of T. b. rhodesiense [60, 62], and they were also documented as reservoirs of T. b. rhodesiense in Kenya [59] and Tanzania [25].
In Malawi, rhodesiense HAT has been a burden for decades [18]. Unlike typical T. b. rhodesiense infections, HAT in Malawi is characterized by the distinct clinical sign of chronic hemolymphatic stage infection without the formation of a chancre; this makes diagnosis difficult [12, 29]. Endemic foci of HAT in the country are the Nkhotakota, Kasungu, and Rumphi districts [11, 30], where large national parks exist. Cattle are one of the most economically important livestock animals in Malawi [10, 48]. A previous study was conducted to update the distribution and clarify the epidemiology of bovine trypanosomiasis caused by T. congolense, T. vivax, and T. brucei in Malawi using an indirect AbELISA serological detection method [58]. However, the specificity of the IgG ELISA was questionable, and as in other serological assays, false positives may have been present [22, 58]. In contrast, PCR of the internal transcribed spacer (ITS) 1 region of ribosomal RNA (rRNA), which can distinguish species by product size, has been widely used to identify trypanosome species [19, 43]. Because the ITS1 region cannot resolve Trypanozoon species to the subspecies level (T. brucei brucei, T. brucei rhodesiense, T. brucei gambiense, T. evansi, and T. equiperdum) [19, 43], detection of T. b. rhodesiense by PCR has been widely conducted by targeting the human serum resistance-associated (SRA) gene [47]. Identifying species by ITS1 PCR is sometimes difficult due to ambiguous and nonspecific signals, which may result in false-positive annotations because of subjective human decisions. In addition, it is impossible to differentiate between T. godfreyi and T. vivax because they share overlapping amplicon size ranges.
MinION, developed by Oxford Nanopore Technologies, is a portable next-generation sequencer (NGS) that connects to a laptop computer through a USB cable [28, 36]. MinION is unique among sequencing tools because it identifies nucleotides in a nanoscale ion channel (nanopore) by detecting specific changes in the electric current when DNA passes through the nanopore [28]. DNA sequencing is a definitive diagnostic method for detecting pathogenic species, and several studies have reported application of MinION for pathogen identification [4, 21, 46, 53]. Unlike conventional sequencers, MinION is economically affordable, allowing sequence analysis without preinstallation of expensive equipment, and not requiring a separate electric supply after connecting to a laptop computer [53]. Further, operating MinION does not require sophisticated skills in biological research [53]. Given these features, genotyping of pathogens on-site with MinION is now feasible [53].
In this study, MinION NGS was combined to PCR-based methods to determine the prevalence of human and animal trypanosomes in cattle and to understand their epidemiological importance for HAT and AAT in Malawi.
Materials and methods
Ethics approval
The ethical clearance for animal sampling was obtained from the Ministry of Agriculture, Irrigation and Water Development in Malawi through the Department of Animal Health and Livestock Development with reference number 10/15/32/D.
Sample collection
This is a descriptive, cross-sectional study of trypanosome infection in cattle, conducted by analyzing DNA material from 446 bovine blood samples. Samples were collected from cattle in three districts in Malawi: Kasungu, Nkhotakota, and Lilongwe, in February and March 2018 during the rainy season. Information on farmers and cattle populations was obtained from the District Agriculture Development Office (DADO) for each district through their District Animal Health and Livestock Development Officer (DAHLDO). The sampling map (Fig. 1) was constructed using the free and open-source geographic information system QGIS [45].
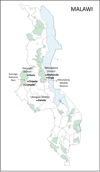 |
Figure 1 Map of Malawi showing sampling locations. Black arrows point to Kasungu National Park and Nkhotakota Wildlife Reserve. Sampling points in Nkhotakota and Kasungu districts were located outside the nature reserves in <1 km proximity. |
Villages located in the vicinity of Kasungu National Park and Nkhotakota Wildlife Reserve were deliberately selected as sampling sites because these areas are locations of human–livestock–wildlife interface and are endemic for tsetse flies carrying T. b. rhodesiense. Both clinically healthy animals and animals with clinical signs were randomly selected within farms. In Kasungu and Nkhotakota districts, a total of 199 and 185 cattle blood samples were collected from 29 and 26 farms, respectively. These farms were selected on purpose from a farmer registry list (Supplementary Table 1), since the selected areas are situated in the human–livestock–wildlife interface area in the vicinity of Kasungu National Park and Nkhotakota Wildlife Reserve (Fig. 1). Both Kasungu National Park and Nkhotakota Wildlife Reserve are tsetse-infested areas and the major HAT/AAT foci in the country [11, 38, 58]. Glossina morsitans morsitans and G. pallidipes are the dominant tsetse fly species in these areas, while G. brevipalpis is also known to exist to some extent [58]. The major cattle breed in these areas is Malawi Zebu, a local breed in the country also known as Angoni cattle (Small East African Zebu) in Eastern Zambia [39]. This breed is known to be susceptible to trypanosomiasis [57]. Cattle are kept in a free-range management system where animals are released during the day to graze freely and return home at dusk. Tsetse flies are often found at the sampling sites; thus, the frequencies of tsetse bite in the cattle in these areas are expected to be high. The other biting flies, tabanids, or Stomoxys spp. are also commonly found in these areas [9, 24].
In Lilongwe district, there are only two commercial farms, and 62 blood samples were collected from one of these farms. Holstein Friesian cattle are maintained with a semi-intensive farm management system, where animals are kept in paddocks. Tsetse flies are not found in this area, and no HAT/AAT cases have been reported in the past from this area [12, 58]. However, the other hematophagous flies including tabanids and Stomoxys spp. are commonly found in Lilongwe, which might facilitate the mechanical transmission of trypanosome, especially T. vivax [44].
Before sampling, the venipuncture site was disinfected with a methylated spirit swab. Then, 5 mL of blood were drawn by venipuncture of the external jugular vein into vacutainer EDTA tubes. Packed cell volume (PCV) counting was conducted to determine anemia; animals with a PCV < 24% were considered anemic. For molecular detection purposes, approximately 200 μL of blood were subjected to DNA extraction using a KURABO QuickGene DNA whole blood kit (Kurabo Industries Ltd.), following the manufacturer’s protocol.
Detection of trypanosomes by ITS1 PCR
DNA samples were subjected to PCR amplification of the rRNA ITS1 region to identify all the pathogenic African trypanosome species/subspecies. ITS1 PCR was carried out using the primers described by Gaithuma et al. [19] (Supplementary Table 2). PCR was performed in a final volume of 25 μL, comprising 12.5 μL Ampdirect plus (Shimadzu, Japan), 0.125 μL BIOTAQ HS DNA Polymerase (5 U/μL) (Bioline, UK), 0.625 μL of each 10 mM primer, 0.5 μL of 2% DMSO, 8.625 μL RNase-free water, and 2 μL extracted DNA. The temperature and cycling profile included initial denaturation at 95 °C for 10 min followed by 37 cycles of denaturation at 94 °C for 30 s, annealing at 53 °C for 90 s, extension at 72 °C for 2 min, and final extension at 72 °C for 7 min. PCR products were examined by electrophoresis in 1.5% agarose S (Nippongene, Japan) in Tris-acetate EDTA (TAE) buffer and stained using GelRed (Biotium, USA) dye before being visualized under ultraviolet light in a transilluminator.
Signals on gel were judged by three investigators independently. They were classified as follows: positive (clear band), negative, and ambiguous (faint band or smearing). Trypanosome species were identified based on the size differences for members of the subgenus Trypanozoon (T. b. brucei, T. evansi, T. b. rhodesiense, and T. b. gambiense), a constant product of approximately 415–431 bp; for T. congolense, 560–705 bp; for T. simiae, 331–343 bp; for T. godfreyi, 220 bp; for T. theileri, 269–350 bp; and for T. vivax, 226–238 bp [19]. The plasmids that contain the TA-cloned ITS1 fragment of T. congolense, T. brucei, T. vivax, T. godfreyi, and T. simiae [40] were combined and used as a positive control for PCR and gel analysis.
MinION library preparation for sequencing
To prepare a MinION sequencing library, amplicons of the ITS1 PCR were used as template then indexes were added by additional PCR. The indexed amplicons were further ligated with adapter DNA provided by Oxford nanopore; then, a sequence-ready library was obtained. Positive, ambiguous samples, and a positive control from ITS1 PCR were sequenced as follows. For the indexing, reagents comprised 5 μL Ampdirect plus (Shimadzu, Japan), 0.05 μL BIOTAQ HS DNA Polymerase (5 U/μL) (Bioline, UK), 0.2 μL of each 10 mM ITS1-index primers (Supplementary Table 2), 0.5 μL DMSO 2%, 2.55 μL RNase-free water, and 2 μL extracted DNA. PCR conditions were as follows: an initial hold at 95 °C for 10 min, followed by 10 cycles of 94 °C for 30 s, 60 °C for 1 min, 72 °C for 2 min, and a final extension at 72 °C for 5 min. Amplicon of the index PCR was prepared for MinION-compatible DNA libraries using a Ligation Sequencing Kit 1D SQK-LSK109 and Native Barcoding Kit EXP-NBD103 (Oxford Nanopore Technologies, UK) as per their instruction manuals. The indexed amplicons were purified by AMPure XP (Beckman Coulter, USA), and equal concentrations of each sample were pooled together to obtain 12 for each pool. End repair and dA-tailing were performed to the pooled, barcoded amplicons using the NEBNext UltraII End Repair/dA-Tailing module (New England Biolabs, UK) per the Oxford Nanopore 1D Native barcoding genomic DNA sequencing protocol. Subsequently, the purified, end-prepped DNA was ligated with the Adapter Mix (AMX) using the NEBNext Quick T4 DNA Ligase (New England Biolabs, UK). Final adapted DNA libraries were purified, and the pre-sequencing mix was eluted and quantified. The flowcell, FLO-MIN107 (Oxford Nanopore Technologies, UK), was primed for loading and sequencing of the library. Before sequencing, MinION and flowcell were assembled and connected to a computer. Platform QC was run using MinKNOW software.
Detection of trypanosome species using MinION sequencing
Base-calling and de-barcoding were conducted using Guppy (Oxford Nanopore Technologies). De-indexing was performed using custom scripts. In brief, indexed primer sequences were aligned on each MinION read using LAST [26], and the best hit indexes at both terminals were subsequently assigned. De-multiplexed reads were aligned with the nucleotide dataset using BLASTn [3]. We kept the best hit for each read and counted the species name that appeared in the output, i.e., T. vivax, T. godfreyi, T. evansi, T. brucei, T. equiperdum, T. congolense, T. simiae, and T. theileri. The counts for T. brucei, T. evansi, and T. equiperdum were summed up and regarded together as Trypanozoon. Each taxon was programmatically assigned on the basis of the population of the read counts if it shared more than 20% of the total and more than 50 read counts. The prevalence of trypanosomes was determined according to the results of MinION sequencing. A schematic workflow of the MinION sequencing and bioinformatic analysis is illustrated in Supplementary Figure.
Detection of T. b. rhodesiense by SRA PCR
SRA PCR was employed to identify T. b. rhodesiense using the primers described by Radwanska et al. [47] (Supplementary Table 2). Reagents used for each reaction included 5 μL Ampdirect plus (Shimadzu, Japan), 0.05 μL BIOTAQ HS DNA Polymerase (5 U/μL) (Bioline, UK), 0.2 μL of each 10 mM primer, 2.55 μL RNase-free water, and 2 μL extracted DNA. The temperature and cycling profile included an initial hold for 10 min at 95 °C, followed by 40 cycles at 94 °C for 30 s, 60 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 5 min. PCR products were examined by electrophoresis in 2% agarose S (Nippongene, Japan) in TAE buffer and stained using GelRed (Biotium, USA) dye before being visualized under ultraviolet light.
Statistical analysis
The chi-square test was used to analyze the relationship between the infection rates by animals, region, and anemic status (p < 0.05).
Results
Trypanosome infections inferred from ITS1 PCR
Forty-four positive, 8 ambiguous, and 394 negative samples were obtained by ITS1 PCR. For positive samples, we identified 21 amplicons with a single band size of 620–700 bp, indicating a single T. congolense infection; 9 amplicons with a single band size of 480 bp, indicating a single Trypanozoon infection; 9 amplicons with a single band size of 250 bp, indicating a single T. vivax or T. godfreyi infection; and 2 amplicons with a single band size of 350 bp, indicating a single T. theileri infection. Three amplicons showed clear multiple bands, indicating two mixed infections of T. congolense and Trypanozoon (A35 and A37; Fig. 2, Supplementary Table 3) and one mixed infection with T. congolense and T. vivax (D66; Fig. 2, Supplementary Table 3).
 |
Figure 2 Gel images of the ITS1 PCR results for positive and ambiguous samples (n = 53, including one positive control). The combination of number and letter referred to the sample identity. M and PC represent the abbreviation of 1 kb marker and positive control, respectively. The letter below explains the interpretation of the gel (Gel) and the analysis of MinION sequencing (Seq). C, T. congolense; T, Trypanozoon; V/G, T. vivax or T. godfreyi; S, T. simiae; Th, T. theileri; N, no result; and “?”, ambiguous result.” |
Trypanosome infections inferred from MinION sequencing
In total, 503,039 reads were obtained from 53 multiplexed ITS1 amplicons, including one positive control. Taxonomic identity was determined for 369,673 reads using a BLAST homology search. For T. congolense, 24 samples (including single and mixed infections) were positive by ITS1 PCR, and T. congolense sequences were detected in the same 24 samples and one additional sample, A17, by the MinION system (Fig. 2, Supplementary Table 3). For T. vivax, 10 samples were positive, and the same 10 samples were confirmed by sequencing, as well as two additional samples, D07 and E60. E60 was a mixed T. congolense and T. vivax infection. For T. theileri, two samples were positive in both ITS1 PCR and the MinION system.
In contrast, 11 samples positive for Trypanozoon were identified by ITS1 PCR. However, using the MinION system, Trypanozoon sequences were detected in only 3 of these 11 products (Supplementary Table 3). Most cases, with the exception of A37, produced reads for Trypanozoon that were much lower than our threshold (50 reads minimum). For A37, 172 reads for Trypanozoon out of 2965 reads were obtained, but reads were below our criteria, with a minimum of 50 reads and more than 20% of total reads; therefore, this sample was negative even though we could not eliminate the possibility of actual Trypanozoon infection.
Three confirmed cases of mixed infections were observed (A35, D66, and E60; Fig. 2, Supplementary Table 3). E60 showed a clear signal for Trypanozoon and a faint signal for T. godfreyi or T. vivax. However, the MinION system provided substantial numbers of sequences to show that the sample was a mixed infection of T. congolense and T. vivax (Supplementary Table 3). Another case, A37, appeared to be a mixed infection of T. congolense and Trypanozoon by ITS1 PCR; however, MinION results did not support Trypanozoon infection as described above.
Prevalence of African trypanosomes determined by ITS1-MinION and SRA-PCR
In addition to ITS1-MinION, SRA PCR was conducted to differentiate T. b. rhodesiense from Trypanozoon. The samples of ITS1-MinION positive for Trypanozoon and SRA-PCR negative were assigned as T. b. brucei. The samples of SRA-PCR positive and ITS1-MinION negative were assigned as T. b. rhodesiense, since SRA PCR has higher sensitivity as discussed later. Trypanosoma congolense was the most prevalent trypanosome detected in Nkhotakota (10.8%; 20 of 185) and Kasungu (2.5%; 5 of 199). Trypanosoma vivax was found in Nkhotakota (6.5%; 12 of 185). Non-pathogenic Trypanosoma species T. theileri (1.1%; 2 of 185) was also detected in Nkhotakota. There were three mixed infections: one T. congolense with T. b. brucei, and two T. congolense with T. vivax (Table 1).
Detection of pathogenic trypanosome species by analysis of SRA PCR and MinION sequencing of ITS1 PCR products.
Trypanosoma brucei rhodesiense was detected in cattle in both HAT-active foci by SRA PCR. A higher prevalence of T. b. rhodesiense was found in samples from Kasungu (9.5%; 19 of 199), followed by Nkhotakota (2.7%; 5 of 185) (Table 1). The detected number of T. b. rhodesiense was much higher than the detected number of Trypanozoon as indicated by the ITS1-MinION detection system. Out of 24 SRA PCR-positive samples, only 1 sample (B23) was positive when analyzed by the ITS1-MinION detection system (Supplementary Table 3).
These trypanosome parasites were all detected in two districts, with the highest prevalence being in Nkhotakota (18.9%; 35 of 185), followed by Kasungu (12.6%; 25 of 199) (Table 1), while no parasites were detected in Lilongwe samples. There was a significant difference in prevalence between the study sites (p < 0.05).
Correlation between infection by African trypanosomes and the anemic status of cattle
There was a significant difference in the anemic status (PCV < 24%) between cattle with trypanosome infection and without infection (p < 0.05) (Table 2). Of 16 anemic cattle, 11 heads (68.7%) were positive for either T. congolense, T. vivax or Trypanozoon (T. b. brucei and T. b. rhodesiense) infections. In contrast, of 430 non-anemic cattle, only 49 heads (11.4%) were positive for trypanosome infection. In particular, infection with T. congolense showed a significant difference in anemia status compared to other species (p < 0.05). A significant difference in the anemic status of T. congolense-infected cattle was observed in both single-infected cattle and mixed-infected cattle (Table 2).
Associations between PCV values and trypanosome species infected in cattle.
Discussion
In this study, we applied next-generation sequencing (NGS) technologies, particularly MinION, a field-friendly, portable, rapid, and affordable NGS device. ITS1 amplicons, including ambiguous signals, were subjected to sequencing analysis. Identification of Trypanosoma sp. by ITS1 PCR and sequencing analysis was largely consistent for T. congolense, T. theileri, and T. vivax. This suggests that MinION sequencing can detect and differentiate trypanosome species. In addition, MinION sequencing appreciably remedied four downsides of ITS1 PCR: ambiguous signals, similar amplicon sizes, nonspecific signals, and multiple infections. We successfully detected a substantial number of reads from samples with ambiguous ITS1 PCR amplicons. T. godfreyi and T. vivax were differentiated. On the other hand, nonspecific amplification hampers correct identification and can lead to false positives. We observed 11 signals of approximately 450 bp corresponding to Trypanozoon; however, most were not supported by sequencing. These could be annotated as false positives without sequencing validation. Apart from nonspecific signals, all samples definitively annotated by the ITS1 PCR had more sequence reads than the threshold of 50. Since we detected enough reads for other trypanosome species from ITS1 PCR, the discrepancy presumably derived from nonspecific PCR amplification of fragments of the same size as the Trypanozoon amplicon. Thus, in several cases, disagreement between ITS1-PCR gel analysis and ITS1-MinION sequence analysis was observed, likely due to nonspecific amplification and misjudgement of the ITS1-PCR gel analysis. Multiple infections were also successfully confirmed by sequencing. All these observations support the assertion that ITS1 PCR validated by sequencing analysis using MinION increases the reliability of parasite detection. Conversely, we obtained only tentative annotation in some samples, even though substantial sequence reads were acquired. Annotation depends on the threshold, and we applied a conservative value to eliminate false positives rather than allow false negatives. BLAST homology search and de-indexing steps were possible sources of error owing to the low accuracy of MinION sequencing. The effective range of ratios and absolute numbers of parasites in mixed infections should also be determined. These points should be clarified and optimized in future studies. The combined approach of ITS1 PCR and MinION has the potential to exclude false positives by nonspecific amplification and less-objective human decisions. Sequencing itself became cost-effective because of the multiplex sequencing system using MinION indexes and barcodes. The quality of field samples is occasionally poor and leads to unexpected nonspecific amplification, as observed in our cases. MinION provides sequence information that can exclude nonspecific amplification and thus improve specificity [65]. Therefore, utilization of MinION-based sequencing in this study will help increase the reliability of PCR-based epidemiology.
ITS1 PCR in combination with MinION sequencing provides sequence information to identify a broad range of trypanosome species in a more reliable manner compared to the conventional ITS1 PCR with gel analysis only [19]. However, this method has a limitation in characterizing Trypanozoon subspecies owing to the highly conserved sequence of the species [14]. In addition, ITS1 PCR is known to have less sensitivity compared to other published primer sets targeting repeat sequences [1, 34, 49], or SRA PCR [51]. In our study, most of the SRA-PCR-positive samples were negative for ITS1 PCR, suggesting that ITS1 PCR alone is of low sensitivity and cannot used to investigate the prevalence of Trypanozoon especially for T. b. rhodesiense as reported before [49]. However, as species-specific primers cannot identify other trypanosomes, the ITS1 PCR system offers value for detecting the parasites broadly in the same reaction with less time and cost. To make the ITS1-MinION system more useful for epidemiological studies, future studies should be required to develop new primers targeting ITS1 or other regions, as a simultaneous sensitive diagnosis method. In the current ITS1-MinION system, the SRA PCR test is still recommended for screening human-infective T. b. rhodesiense in a more sensitive and specific manner.
In this study, the most common trypanosome detected was T. congolense. This is in agreement with reports from other southern African countries [32, 58]. Trypanosoma congolense and T. vivax were more prevalent in Nkhotakota than in Kasungu, with no parasites seen in Lilongwe. Possible reasons for this trend may be the different methods applied for AAT control. Nkhotakota wildlife services have intensified blue cloth targets in the park to reduce the number of tsetse flies. In Kasungu, local veterinarians and farmers apply an anti-parasitic treatment, Berenil (diminazene aceturate), to the cattle twice every year before and after the herding season. Berenil is a common therapeutic and preventive medicine for animal trypanosomiasis that has been used for >60 years [27]. Since samples were collected during the rainy season, we may expect a higher prevalence of parasites in the dry season when disease transmission peaks [31, 42]. In both the Kasungu and Nkhotakota areas, cattle are usually bitten when brought into close contact with flies at river crossings, village water holes, or other tsetse fly habitats in the field [37]. On the other hand, semi-intensive farming was applied in Lilongwe, where the cattle were kept in paddocks, thus preventing the mechanical transmission of T. vivax in cattle by biting flies. However, as T. vivax was detected in Nkhotakota, this emphasizes the necessity of sustained non-tsetse vector control for animal trypanosomiasis in the region. Also, since we collected a limited number of cattle samples only at three locations during short period, a broader countrywide survey is required to assess the overall AAT/HAT situation in the country.
The PCV value of infected cattle was significantly lower than that of non-infected cattle. Anemia has long been considered the main clinical sign of trypanosomiasis in both humans and animals [15, 16]. Anemia associated with infection has been previously associated with lower productivity of infected animals [55, 56]. Consistent with previous studies [33, 54], our findings confirmed that cattle with T. congolense infection tended to be anemic, as compared to those infected with other trypanosome species, suggesting the importance of controlling AAT.
Malawi is rich in ecosystems where humans, livestock, and wildlife populations exist close to each other. Compared with neighboring countries such as Uganda, Kenya, and Tanzania in eastern and southern Africa where rhodesiense HAT is endemic [5, 59, 62], T. b. rhodesiense prevalence in tsetse flies is higher in Malawi, according to xenomonitoring data [2]. As Kasungu and Nkhotakota are hotspots of HAT in Malawi, the presence of T. b. rhodesiense in cattle has been suspected in these areas [11, 38], but had not been investigated before this study.
Studying the human–livestock–wildlife interface is essential now, and even more so in the future because of human population expansion and agricultural development [20]. This expansion induces farmers and their livestock to migrate closer to wildlife conservation areas, increasing their exposure to tsetse flies. Here, the surveyed areas were in close proximity, within <1 km, to the Kasungu and Nkhotakota national parks. The extensive farming system allowed cattle to interact with infected wildlife and tsetse flies during grazing activities at the human–livestock–wildlife interface, increasing the possibility of pathogen sharing and disease transmission in the populations involved.
In this study, we identified both human- and animal-infective trypanosomes residing in cattle at the human–livestock–wildlife interface areas in Malawi. A limited number of trypanosomiasis and tsetse control programs addressing both diseases have been conducted in the area [35]. AAT and HAT control activities are interdependent since both diseases share the same transmission vector and host. Control programs targeting flies and animal populations are necessary to achieve HAT control [23]. This study contributes to improving knowledge regarding the status of trypanosomiasis in Malawi. Our findings emphasize the need for sustainable integration between AAT and HAT control measures and collaborative human and animal health care services under the One Health concept, which are indispensable in tackling HAT.
Conclusions
This is the first study to assess the prevalence of animal and human infective trypanosomes in cattle in Malawi. The use of MinION sequencing in combination with ITS1 PCR increased the reliability of PCR-based comprehensive trypanosome detection. However, for more sensitive and specific detection of human infective T. b. rhodesiense, SRA PCR is still recommended. Trypanosome-susceptible cattle harbor both human and animal infective trypanosomes, implying its role as a potential reservoir of T. b. rhodesiense. This study emphasizes the urgent need for sustainable control measures within the context of the One Health approach for AAT and HAT in livestock.
Supplementary materials
Supplementary Table 1
Farmer registry list used during blood samples collection in the districts of Kasungu and Nkhotakota. (*) indicates the data obtained from the District Agriculture Development Office (DADO) for each district through their District Animal Health and Livestock Development Officer (DAHLDO). Access here
Supplementary Table 2
List of primers and their sequences used in this study. Access here
Supplementary Table 3
SRA PCR, ITS PCR, and MinION sequencing analysis for characterizations of trypanosome species. No hit* shows the number of reads that could not be identified by BLAST. FASTA $ describes the total number of obtained reads processed into BLAST. “PC+” and “?” refer to positive control and ambiguous results, respectively. Samples which were negative in both the SRA PCR and the ITS1 PCR are not shown here. Access here
Supplementary Figure
Workflow for molecular detection of African trypanosomes. The workflow depicts structural steps for detection of African animal and human trypanosomes with serial arrows to the right for cattle samples, and a single downward arrow for human samples. The section on the identification of trypanosome species by sequencing is divided into three steps: library preparation, sequencing and basecalling, and de-multiplexing. Bioinformatic analyses required in the experiments are framed in red borderline. Access hereConflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This study was supported by the Japan Program for Infectious Diseases Research and Infrastructure from the Japan Agency for Medical Research and Development (AMED) under grant number JP20wm0125008 and the International Collaborative Research Program for Tackling NTD (Neglected Tropical Disease) Challenges in African countries under grant number JP18jm0510001. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Ahmed HA, Picozzi K, Welburn SC, MacLeod ET. 2013. A comparative evaluation of PCR-based methods for species-specific determination of African animal trypanosomes in Ugandan cattle. Parasites & Vectors, 6(1), 316. [CrossRef] [PubMed] [Google Scholar]
- Alibu VP, John CK, Matovu E, Malele II, Chisi JE, Mbongo N, Mansinsa P, Intisar ER, Mohammed Y, Abdelrahman MM, Ochi EB, Lukaw YS. 2015. Molecular xenomonitoring of trypanosomes in tsetse flies. Journal of Parasitology and Vector Biology, 7(6), 108–114. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. [CrossRef] [PubMed] [Google Scholar]
- Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, Wain J, O’grady J.. 2014. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nature Biotechnology, 33(3), 296–300. [Google Scholar]
- Auty HK, Picozzi K, Malele I, Torr SJ, Cleaveland S, Welburn S. 2012. Using molecular data for epidemiological inference: assessing the prevalence of Trypanosoma brucei rhodesiense in tsetse in Serengeti, Tanzania. PLoS Neglected Tropical Diseases, 6(1), e1501. [CrossRef] [PubMed] [Google Scholar]
- Bekele EE, Begejo B. 2015. The current situation and diagnostic approach of Nagana in Africa: a review. Journal of Natural Sciences Research, 5(17), 117–125. [Google Scholar]
- Brun R, Blum J, Chappuis F, Burri C. 2010. Human African trypanosomiasis. Lancet, 375(9709), 148–159. [CrossRef] [PubMed] [Google Scholar]
- Büscher P, Cecchi G, Jamonneau V, Priotto G. 2017. Human African trypanosomiasis. Lancet, 390(10110), 2397–2409. [CrossRef] [PubMed] [Google Scholar]
- Chainey JE, Oldroyd H. 1980. Family Tabanidae. pp. 275–308, in Catalogue of the Diptera of the Afrotropical Region, Crosskey R.W., Editor. British Museum (Natural History): London. 1437 p. [Google Scholar]
- Chintsanya NC, Chinombo DO, Gondwe TN, Wanda G, Mwenda AR, Banda MC, Hami JC. 2004. Management of farm animal genetic resources in the Sadc Region-Malawi. Available from: http://www.fao.org/tempref/docrep/fao/011/a1250f/annexes/CountryReports/Malawi.pdf. [Google Scholar]
- Chisi JE, Muula AS, Ngwira B, Kabuluzi S. 2011a. A retrospective study of human African Trypanosomiasis in three Malawian districts. Tanzania Journal of Health Research, 13(1), 62–68. [Google Scholar]
- Chisi J, Nkhoma A, Sternberg J. 2011b. Presentation of Trypanosomiasis in Nkhotakota. Malawi Medical Journal, 19(4), 140–141. [Google Scholar]
- Connor RJ. 1994. The impact of nagana. Onderstepoort Journal of Veterinary Research, 61, 379–383. [Google Scholar]
- Cuypers B, Van den Broeck F, Van Reet N, Meehan CJ, Cauchard J, Wilkes JM, Claes F, Goddeeris B, Birhanu H, Dujardin JC, Laukens K. 2017. Genome-wide SNP analysis reveals distinct origins of Trypanosoma evansi and Trypanosoma equiperdum. Genome Biology and Evolution, 9(8), 1990–1997. [CrossRef] [PubMed] [Google Scholar]
- Fiennes RN. 1954. Haematological studies in trypanosomiasis of cattle. Veterinary Record, 66(30), 423–434. [Google Scholar]
- Fiennes RN. 1970. Pathogenesis and pathology of animal trypanosomiasis, in The African Trypanosomiasis, Mulligan HW, Editor. Allen and Unwin: London. pp. 729–750. [Google Scholar]
- Franco JR, Simarro PP, Diarra A, Jannin JG. 2014. Epidemiology of human African trypanosomiasis. Clinical Epidemiology, 6, 257. [PubMed] [Google Scholar]
- Franco JR, Cecchi G, Priotto G, Paone M, Diarra A, Grout L, Mattioli RC, Argaw D. 2017. Monitoring the elimination of human African trypanosomiasis: update to 2014. PLoS Neglected Tropical Diseases, 11(5), e0005585. [CrossRef] [PubMed] [Google Scholar]
- Gaithuma AK, Yamagishi J, Martinelli A, Hayashida K, Kawai N, Marsela M, Sugimoto C. 2019. A single test approach for accurate and sensitive detection and taxonomic characterization of trypanosomes by comprehensive analysis of internal transcribed spacer 1 amplicons. PLoS Neglected Tropical Diseases, 13(2), e0006842. [CrossRef] [PubMed] [Google Scholar]
- Gondwe N, Marcotty T, Vanwambeke SO, De Pus C, Mulumba M, Van den Bossche P. 2009. Distribution and density of tsetse flies (Glossinidae: Diptera) at the game/people/livestock interface of the Nkhotakota Game Reserve human sleeping sickness focus in Malawi. EcoHealth, 6(2), 260–265. [CrossRef] [PubMed] [Google Scholar]
- Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, Stryke D, Bouquet J, Somasekar S, Linnen JM, Dodd R. 2015. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Medicine, 7(1), 99. [CrossRef] [PubMed] [Google Scholar]
- Hopkins JS, Chitambo H, Machila N, Luckins AG, Rae PF, van den Bossche P, Eisler MC. 1998. Adaptation and validation of antibody-ELISA using dried blood spots on filter paper for epidemiological surveys of tsetse-transmitted trypanosomosis in cattle. Preventive Veterinary Medicine, 37(1–4), 91–99. [CrossRef] [PubMed] [Google Scholar]
- Jali K. 2019. Trypanosomiasis (African) – Malawi (02): (Nkhotakota Wildlife Reserve). ProMed. 01 Nov: 20191101.6757106. http://www.promedmail.org. Accessed 2 November 2019. [Google Scholar]
- Ježek J, Votýpka J, Brzoňová J, Oboňa J. 2019. Horse flies (Diptera: Tabanidae) collected in Central African Republic, Gabon and Liberia with comments on their updated distribution. Acta Musei Silesiae, Scientiae Naturales, 68(3), 263–274. [CrossRef] [Google Scholar]
- Kaare MT, Picozzi K, Mlengeya T, Fèvre EM, Mellau LS, Mtambo MM, Cleaveland S, Welburn SC. 2007. Sleeping sickness-A re-emerging disease in the Serengeti? Travel Medicine and Infectious Disease, 5(2), 117–124. [CrossRef] [PubMed] [Google Scholar]
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Research, 21(3), 487–493. [CrossRef] [PubMed] [Google Scholar]
- Kuriakose SM, Onyilagha C, Singh R, Jia P, Uzonna JE. 2019. Diminazene aceturate (Berenil) downregulates Trypanosoma congolense-induced proinflammatory cytokine production by altering phosphorylation of MAPK and STAT proteins. Immunologic Research, 67(1), 84–92. [CrossRef] [PubMed] [Google Scholar]
- Loman NJ, Watson M. 2015. Successful test launch for nanopore sequencing. Nature Methods, 12, 303–304. [CrossRef] [PubMed] [Google Scholar]
- MacLean LM, Odiit M, Chisi JE, Kennedy PG, Sternberg JM. 2010. Focus specific clinical profiles in human African trypanosomiasis caused by Trypanosoma brucei rhodesiense. PLoS Neglected Tropical Diseases, 4(12), e906. [CrossRef] [PubMed] [Google Scholar]
- Madanitsa M, Chisi J, Ngwira B. 2009. The epidemiology of trypanosomiasis in Rumphi district, Malawi: a ten year retrospective study. Malawi Medical Journal, 21(1), 22–27. [CrossRef] [Google Scholar]
- Majekodunmi AO, Fajinmi A, Dongkum C, Picozzi K, MacLeod E, Thrusfield MV, Shaw AP, Welburn SC. 2013. Social factors affecting seasonal variation in bovine trypanosomiasis on the Jos Plateau, Nigeria. Parasites & Vectors, 6(1), 293. [CrossRef] [PubMed] [Google Scholar]
- Mamabolo MV, Ntantiso L, Latif A, Majiwa PA. 2009. Natural infection of cattle and tsetse flies in South Africa with two genotypic groups of Trypanosoma congolense. Parasitology, 136(4), 425–431. [CrossRef] [PubMed] [Google Scholar]
- Mamoudou A, Payne VK, Sevidzem SL. 2015. Hematocrit alterations and its effects in naturally infected indigenous cattle breeds due to Trypanosoma spp. on the Adamawa Plateau-Cameroon. Veterinary World, 8(6), 813. [CrossRef] [PubMed] [Google Scholar]
- Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. 1992. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. International Journal for Parasitology, 22(7), 909–918. [CrossRef] [PubMed] [Google Scholar]
- Meyer A, Holt HR, Selby R, Guitian J. 2016. Past and ongoing tsetse and animal trypanosomiasis control operations in five African countries: a systematic review. PLoS Neglected Tropical Diseases, 10(12), e0005247. [CrossRef] [PubMed] [Google Scholar]
- Mongan AE, Tuda JS, Runtuwene LR. 2020. Portable sequencer in the fight against infectious disease. Journal of Human Genetics, 65, 35–40. [CrossRef] [PubMed] [Google Scholar]
- Mulligan HW, Porrs W. 1970. The African trypanosomiases. New York: Wiley-Interscience. 325 p. [Google Scholar]
- Musaya J, Chisi J, Senga E, Nambala P, Maganga E, Matovu E, Enyaru J. 2017. Polymerase chain reaction identification of Trypanosoma brucei rhodesiense in wild tsetse flies from Nkhotakota Wildlife Reserve, Malawi. Malawi Medical Journal, 29(1), 11–15. [CrossRef] [Google Scholar]
- Mwai O, Hanotte O, Kwon YJ, Cho S. 2015. African indigenous cattle: unique genetic resources in a rapidly changing world. Asian-Australasian Journal of Animal Sciences, 28(7), 911. [CrossRef] [PubMed] [Google Scholar]
- Nambala P, Musaya J, Hayashida K, Maganga E, Senga E, Kamoto K, Chisi J, Sugimoto C. 2018. Comparative evaluation of dry and liquid RIME LAMP in detecting trypanosomes in dead tsetse flies. Onderstepoort Journal of Veterinary Research, 85(1), 1–6. [CrossRef] [Google Scholar]
- Ng’ayo MO, Njiru ZK, Kenya EU, Muluvi GM, Osir EO, Masiga DK. 2005. Detection of trypanosomes in small ruminants and pigs in western Kenya: important reservoirs in the epidemiology of sleeping sickness? Kinetoplastid Biology and Disease, 4(1), 5. [CrossRef] [PubMed] [Google Scholar]
- Ngonyoka A, Gwakisa PS, Estes AB, Salekwa LP, Nnko HJ, Hudson PJ, Cattadori IM. 2017. Patterns of tsetse abundance and trypanosome infection rates among habitats of surveyed villages in Maasai steppe of northern Tanzania. Infectious Diseases of Poverty, 6(1), 126. [CrossRef] [PubMed] [Google Scholar]
- Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, Dávila AM. 2005. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitology Research, 95(3), 186–192. [CrossRef] [PubMed] [Google Scholar]
- Osório AL, Madruga CR, Desquesnes M, Soares CO, Ribeiro LR, Costa SC. 2008. Trypanosoma (Duttonella) vivax: its biology, epidemiology, pathogenesis, and introduction in the New World – a review. Memórias do Instituto Oswaldo Cruz, 103(1), 1–13. [CrossRef] [Google Scholar]
- QGIS Development Team. 2020. QGIS Geographic Information System. Open Source Geospatial Foundation Project. http://qgis.osgeo.org. [Google Scholar]
- Quick J, Quinlan AR, Loman NJ. 2014. A reference bacterial genome dataset generated on the MinION portable single-molecule nanopore sequencer. Gigascience, 3, 1–6. [CrossRef] [PubMed] [Google Scholar]
- Radwanska M, Chamekh M, Vanhamme LU, Claes F, Magez S, Magnus E, De Baetselier P, Büscher P, Pays E. 2002. The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. American Journal of Tropical Medicine and Hygiene, 67(6), 684–690. [CrossRef] [Google Scholar]
- Schmidt RH. 1969. Cattle in Malawi’s Southern region. Society of Malawi Journal, 22, 57–72. [Google Scholar]
- Sloof P, Menke HH, Caspers MP, Borst P. 1983. Size fractionation of Trypanosoma brucei DNA: localization of the 177-bp repeat satellite DNA and a variant surface glycoprotein gene in a mini-chromosomal DNA fraction. Nucleic Acids Research, 11(12), 3889–3901. [CrossRef] [PubMed] [Google Scholar]
- Spickler AR. 2018. African Animal Trypanosomiasis. Retrieved from: http://www.cfsph.iastate.edu/Factsheets/pdfs/trypanosomiasis_african.pdf. [Google Scholar]
- Squarre D, Hayashida K, Gaithuma A, Chambaro H, Kawai N, Moonga L, Namangala B, Sugimoto C, Yamagishi J. 2020. Diversity of trypanosomes in wildlife of the Kafue ecosystem, Zambia. International Journal for Parasitology: Parasites and Wildlife, 12, 34–41. [CrossRef] [Google Scholar]
- Steverding D. 2008. The history of African trypanosomiasis. Parasites & Vectors, 1(1), 3. [CrossRef] [PubMed] [Google Scholar]
- Sugano S, Yamagishi J, Mongan AE, Tuda J, Suzuki Y. 2015. Genomics on site of detection of malaria. Annals of Translational Medicine, 3(S2), AB005. [Google Scholar]
- Tasew S, Duguma R. 2012. Cattle anaemia and trypanosomiasis in western Oromia State, Ethiopia. Revue de Médecine Véterinaire, 163(12), 581–588. [Google Scholar]
- Trail JC, d’Ieteren GD, Feron A, Kakiese O, Mulungo M, Pelo M. 1990. Effect of trypanosome infection, control of parasitaemia and control of anaemia development on productivity of N’Dama cattle. Acta Tropica, 48(1), 37–45. [CrossRef] [PubMed] [Google Scholar]
- Trail JC, d’Ieteren GD, Murray M, Ordner G, Yangari G, Collardelle C, Sauveroche B, Maille JC, Viviani P. 1993. Measurement of trypanotolerance criteria and their effect on reproductive performance of N’Dama cattle. Veterinary Parasitology, 45(3–4), 241–255. [CrossRef] [PubMed] [Google Scholar]
- Van den Bossche PR, Rowlands GJ. 2001. The relationship between the parasitological prevalence of trypanosomal infections in cattle and herd average packed cell volume. Acta Tropica, 78(2), 163–170. [CrossRef] [PubMed] [Google Scholar]
- Van den Bossche P, Shumba W, Makhambera P. 2000. The distribution and epidemiology of bovine trypanosomosis in Malawi. Veterinary Parasitology, 88(3–4), 163–176. [CrossRef] [PubMed] [Google Scholar]
- Von Wissmann B, Machila N, Picozzi K, Fèvre EM, Barend M, Handel IG, Welburn SC. 2011. Factors associated with acquisition of human infective and animal infective trypanosome infections in domestic livestock in western Kenya. PLoS Neglected Tropical Diseases, 5(1), e941. [CrossRef] [PubMed] [Google Scholar]
- Von Wissmann B, Fyfe J, Picozzi K, Hamill L, Waiswa C, Welburn SC. 2014. Quantifying the association between bovine and human trypanosomiasis in newly affected sleeping sickness areas of Uganda. PLoS Neglected Tropical Diseases, 8(6), e2931. [CrossRef] [PubMed] [Google Scholar]
- Waiswa C, Olaho-Mukani W, Katunguka-Rwakishaya E. 2003. Domestic animals as reservoirs for sleeping sickness in three endemic foci in south–eastern Uganda. Annals of Tropical Medicine & Parasitology, 97(2), 149–155. [CrossRef] [Google Scholar]
- Welburn SC, Picozzi K, Fèvre EM, Coleman PG, Odiit M, Carrington M, Maudlin I. 2001. Identification of human-infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet, 358(9298), 2017–2019. [CrossRef] [PubMed] [Google Scholar]
- W.H.O. World Health Organization. 2013. Control and surveillance of human African trypanosomiasis report of a WHO Expert Committee WHO Technical Report Series 984. Geneva, Switzerland: World Health Organization. [Google Scholar]
- W.H.O. World Health Organization. 2019. Trypanosomiasis, human African (sleeping sickness). https://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness). Accessed 20 October 2019. [Google Scholar]
- Yamagishi J, Runtuwene LR, Hayashida K, Mongan AE, Thi LA, Thuy LN, Nhat CN, Limkittikul K, Sirivichayakul C, Sathirapongsasuti N, Frith M. 2017. Serotyping dengue virus with isothermal amplification and a portable sequencer. Scientific Reports, 7(1), 1–10. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Marsela M, Hayashida K, Nakao R, Chatanga E, Gaithuma AK, Naoko K, Musaya J, Sugimoto C & Yamagishi J. 2020. Molecular identification of trypanosomes in cattle in Malawi using PCR methods and nanopore sequencing: epidemiological implications for the control of human and animal trypanosomiases. Parasite 27, 46.
All Tables
Detection of pathogenic trypanosome species by analysis of SRA PCR and MinION sequencing of ITS1 PCR products.
All Figures
 |
Figure 1 Map of Malawi showing sampling locations. Black arrows point to Kasungu National Park and Nkhotakota Wildlife Reserve. Sampling points in Nkhotakota and Kasungu districts were located outside the nature reserves in <1 km proximity. |
| In the text | |
 |
Figure 2 Gel images of the ITS1 PCR results for positive and ambiguous samples (n = 53, including one positive control). The combination of number and letter referred to the sample identity. M and PC represent the abbreviation of 1 kb marker and positive control, respectively. The letter below explains the interpretation of the gel (Gel) and the analysis of MinION sequencing (Seq). C, T. congolense; T, Trypanozoon; V/G, T. vivax or T. godfreyi; S, T. simiae; Th, T. theileri; N, no result; and “?”, ambiguous result.” |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


