| Issue |
Parasite
Volume 27, 2020
|
|
|---|---|---|
| Article Number | 39 | |
| Number of page(s) | 9 | |
| DOI | https://doi.org/10.1051/parasite/2020033 | |
| Published online | 26 May 2020 | |
Research Article
A novel nuclear marker and development of an ARMS-PCR assay targeting the metallopeptidase 10 (nas 10) locus to identify the species of the Anisakis simplex (s. l.) complex (Nematoda, Anisakidae)
Un nouveau marqueur nucléaire et développement d'un test ARMS-PCR ciblant le locus de la métallopeptidase 10 (nas 10) pour identifier les espèces du complexe Anisakis simplex (s. l.) (Nematoda, Anisakidae)
1
Department of Public Health and Infectious Diseases, Section of Parasitology, “Sapienza – University of Rome”, Piazzale Aldo Moro 5, 00185 Rome, Italy
2
Department of Ecological and Biological Sciences, Tuscia University, Viale dell’Università s/n, 01100 Viterbo, Italy
3
Cawthron Institute, 98 Halifax Street East, Nelson 7010, New Zealand
* Corresponding Author: Simonetta.mattiucci@uniroma1.it
Received:
10
February
2020
Accepted:
30
April
2020
The genus Anisakis represents one of the most widespread groups of ascaridoid nematodes in the marine ecosystem. Three closely related taxa are recognized in the Anisakis simplex (s. l.) complex: A. pegreffii, A. simplex (s. s.) and A. berlandi. They are widely distributed in populations of their intermediate/paratenic hosts (fish and squids) and definitive hosts (cetaceans). A novel nuclear gene locus, metallopeptidase 10 (nas 10) (451 bp), was sequenced and validated on a total of 219 specimens of the three species of Anisakis, collected in fish and cetacean hosts from allopatric areas included in their ranges of distribution. The specimens of Anisakis were first identified by allozymes and sequence analysis of the mtDNA cox2 and EF1α-1 nDNA. The novel nuclear marker has shown fixed alternative nucleotide positions in the three species, i.e. diagnostic at 100%, permitting the species determination of a large number of specimens analyzed in the present study. In addition, primers to be used for amplification-refractory mutation system (ARMS) PCR of the same gene locus were designed at these nucleotide positions. Thus, direct genotyping determination, by double ARMS, was developed and validated on 219 specimens belonging to the three species. Complete concordance was observed between the tetra-primer ARMS-PCR assays and direct sequencing results obtained for the nas 10 gene locus. The novel nuclear diagnostic marker will be useful in future studies on a multi-locus genotyping approach and also to study possible hybridization and/or introgression events occurring between the three species in sympatric areas.
Résumé
Le genre Anisakis représente l’un des groupes de nématodes ascaridoïdes les plus répandus dans l’écosystème marin. Trois taxons étroitement apparentés sont reconnus dans le complexe Anisakis simplex (s. l.) : A. pegreffii, A. simplex (s. s.) et A. berlandi. Ils sont largement répartis dans les populations de leurs hôtes intermédiaires/paraténiques (poissons et calmars) et définitifs (cétacés). Un nouveau locus de gène nucléaire, la métallopeptidase 10 (nas 10) (451 pb), a été séquencé et validé sur un total de 219 spécimens des trois espèces d’Anisakis, collectés chez des hôtes poissons et cétacés de zones allopatriques incluses dans leur aire de répartition. Les échantillons d’Anisakis ont d’abord été identifiés par des allozymes et une analyse des séquences de l’ADNmt cox2 et de l’ADNn EF1α-1. Le nouveau marqueur nucléaire a montré des positions de nucléotides alternatives fixes dans les trois espèces, c’est-à-dire qu’il a permis un diagnostic à 100%, permettant la détermination de l’espèce d’un grand nombre d’échantillons analysés dans la présente étude. De plus, des amorces à utiliser pour la PCR par système de mutation réfractaire à l’amplification (ARMS) du même locus génique ont été conçues à ces positions nucléotidiques. Ainsi, la détermination directe du génotypage, par double ARMS, a été développée et validée sur 219 spécimens appartenant aux trois espèces. Une concordance complète a été observée entre les dosages ARMS PCR tétra-amorces et les résultats de séquençage direct obtenus pour le locus du gène nas 10. Le nouveau marqueur de diagnostic nucléaire sera utile dans les travaux futurs d’une approche de génotypage multi-locus et également pour étudier les éventuels événements d’hybridation et/ou d’introgression se produisant entre les trois espèces dans des zones sympatriques.
Key words: Anisakis simplex (s. s.) / A. pegreffii / A. berlandi / Diagnostic markers / Metallopeptidase nas 10 nDNA / ARMS-PCR
© M. Palomba et al., published by EDP Sciences, 2020
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The Anisakis simplex (s. l.) species complex includes heteroxenous nematodes belonging to three distinct species, namely A. pegreffii, A. simplex (s. s.) and A. berlandi [25]. The three taxa, first genetically detected by allozymes [20], were later morphologically described [23], and recognized as distinct biological species based on different nuclear [33] and mitochondrial gene loci [35]. The geographical range of these three species is quite distinct; however, some oceanographic basin waters are reported as overlapping areas and contact zones between them (for a review, [25]). In allopatric and sympatric areas, they are known to infect several definitive and intermediate/paratenic hosts; the three species often being found parasitizing the same host-species range [25]. In addition, A. pegreffii and A. simplex (s. s.) are recognized as zoonotic parasites that cause human gastro- (GA) intestinal- (IA) anisakiasis and gastro-allergic (GAA) anisakiasis [25]. Gastro-intestinal anisakiasis is currently widespread in Europe, mostly occurring in Italy, Spain, France, Portugal and Croatia (reviewed in [9, 25]); cases have also been widely reported in Japan and South Korea [15]. Humans are accidental hosts who become infected after the ingestion of raw fish and squids containing, in their edible parts, the third-stage larvae (L3) of A. pegreffii and A. simplex (s. s.). No data are so far available about the zoonotic role of the species A. berlandi. On the other hand, while it was widely demonstrated that A. simplex (s. s.) and A. pegreffii [6, 13, 14] infect the musculature of the fish hosts, there is no knowledge about the ability of A. berlandi to invade the fish host musculature.
In recent years, the multi-locus genotyping approach has been suggested to identify nematodes of the genus Anisakis and to investigate certain micro-evolutionary aspects, such as hybridization and introgression phenomena [25]. With this aim, there have been several recent attempts to identify new nuclear markers. These approaches include: (1) sequence analysis of the nuclear locus EF1 α-1 nDNA showing the presence of fixed alternative nucleotide positions between the two species A. pegreffii and A. simplex (s. s.) [24], and (2) the development of DNA microsatellite loci [27]. According to the findings of these authors, no diagnostic markers were discovered between the two taxa [27]. On the other hand, opposite results were obtained by further recent development of polymorphic SSRs DNA markers [26]. Diagnostic SSRs DNA loci between the species A. pegreffii and A. simplex (s. s.) were detected [26], also with respect to the species A. berlandi [3], allowing the reliable identification of the species. It was also found that among the diagnostic loci detected at the SSRs loci between the three Anisakis species [3, 26], sex-linked loci exist. This last finding, however, highlighted the problem that, because male worms are hemizygotes at these diagnostic SSRs loci, their genotypes cannot be considered in a large genotyping analysis in the investigation of hybridization and introgression phenomena [26]. Moreover, it has also been underlined that a multi-locus nuclear genotyping approach is generally mandatory and highly recommended to investigate the occurrence of mixed ancestry only in the hosts and geographic areas where parasite species overlap in their range of distribution [1, 2, 7, 25]. Therefore, the discovery of further nuclear diagnostic markers to be used for disentangling hybridization and/or introgression events between these sibling species of Anisakis is highly desirable.
The amplification-refractory mutation system (ARMS) can distinguish a single base sequence difference using one-step PCR [32]. This method is based on the use of sequence-specific PCR primers that allow amplification of test DNA only when the target allele is contained within the sample. Following an ARMS reaction, the presence or absence of a PCR product is diagnostic for the presence or absence of the target allele. An ARMS-PCR assay was recently developed on ITS rDNA for the species identification of A. pegreffii, A. simplex (s. s.) and A. typica occurring in fish from Korean waters [11].
The scope of this work was (1) to provide a new nuclear diagnostic marker for the three Anisakis species (i.e. A. pegreffii, A. simplex (s. s.) and A. berlandi), based on the direct sequence analysis of a gene locus coding for a metallopeptidase 10 (nas 10), to be used in a future multi-locus approach; (2) to validate the novel marker on a large number of specimens of the three Anisakis species; and (3) to develop and validate an ARMS-PCR tool to genotype the diagnostic single nucleotide polymorphisms (SNPs) detected at nas 10, for rapid genotyping of individuals of A. simplex (s. s.), A. pegreffii and A. berlandi.
Materials and methods
Parasite sampling and molecular identification
A total of 219 Anisakis spp. third-stage larvae and adults collected from intermediate/paratenic fish and cetacean hosts, respectively were obtained from the frozen and alcohol collection stored in the Section of Parasitology, Department of Public Health and Infectious Diseases of “Sapienza – University in Rome”. Details concerning the sampling localities are given in Table 1.
Sampling area, host species and life-history stage (A: adult; L4: 4th stage larvae; L3: 3rd stage larvae) of the specimens of A. simplex (s. s.), A. pegreffii and A. berlandi analyzed.
The allozymes proven to be diagnostic for the species of the A. simplex (s. l.) complex were used [22, 23]. Standard horizontal starch gel electrophoresis was performed at these enzyme loci; their staining procedures are those previously reported [23]. Total DNA was extracted from a tissue portion (≈2 mg) of each Anisakis larva (L3, L4) and each adult specimen (Table 1). The Quick-gDNATM Miniprep Kit (ZYMO RESEARCH) was used as the extraction method. DNA obtained was quantified using a NanoDrop®TC1-E20 spectrophotometer (BioTek Synergy HT).
The specimens were identified to the species level by sequences analysis of the mtDNA cox2 gene locus, which makes it possible to distinguish the three species A. simplex (s. s.), A. pegreffii, and A. berlandi [23]. Additionally, the specimens recognized at the mtDNA cox2 locus as belonging to A. pegreffii and A. simplex (s. s.) were also identified by sequences analysis of EF1 α-1 nDNA [24].
For sequencing of the mtDNA cox2 gene locus, PCR amplification was performed using the primers 211F (5′–TTT TCT AGT TAT ATA GAT TGR TTT YAT–3′) and 210R (5′–CAC CAA CTC TTA AAA TTA TC–3′) [23, 35]. PCR conditions were established as previously described [24]. The amplification of EF1 α-1 nDNA was performed using the primers EF-F (5′–TCC TCA AGC GTT GTT ATC TGT T–3′) and EF-R (5′–AGT TTT GCC ACT AGC GGT TCC–3′), under the conditions previously described [24].
Primer selection for the nas 10 nDNA and PCR condition
For direct sequencing of the metallopeptidase (nas 10 nDNA) gene locus of Anisakis, oligonucleotide primers were newly designed based on the sequence at the metallopeptidase asnas 10 of Ascaris suum, deposited in GenBank under accession number JI170268.1, by using the software program PRIMER3 plus (http://bioinfo.ut.ee/primer3-0.4.0/). The total sample of N = 219 individuals, belonging to the three studied species, previously identified by allozymes, mtDNA cox2 and EF1 α-1 nDNA, was thus sequenced at the metallopeptidase 10 (nas 10) nDNA gene locus. Thus, PCR amplification was performed using the primers nas10-F (5′–GAT GTT CCT GCA AGT GAT TG–3′) and nas10-R (5′–CGC TAT TAA GAG AGG GAT CG–3′) (Table 2). PCR reactions were carried out in a 25 μL volume containing 0.6 μL of each primer 10 mm, 2 μL of MgCl2 25 mm (Promega), 5 μL of 5× buffer (Promega), 0.6 μL of dNTPs 10 mm (Promega), 0.2 μL of Go-Taq Polymerase (5U/μL) (Promega) and 2 μL of total DNA. PCR temperature conditions were the following: 94 °C for 5 min (initial denaturation), followed by 40 cycles at 95 °C for 1 min (denaturation), 53 °C for 1 min (annealing), 72 °C for 1 min (extension), and post-amplification at 72 °C for 15 min.
Primer sequences targeting the metallopeptidase 10 (nas 10) locus, product size, and annealing temperature (T a/°C). In bold, a deliberate second mismatch.
Sequence data analysis
Clustal X (version 2.0) software [12] was used for sequence alignment, obtained at the studied gene loci (i.e. mtDNA cox2, EF1 α-1 nDNA and nas 10 nDNA). In the case of mtDNA cox2 and EF1 α-1 nDNA, the generated sequences were aligned with those from the same species previously obtained [22, 23], to detect the fixed diagnostic nucleotide positions that can be used to discriminate A. simplex (s. s.), A. pegreffii and A. berlandi [22, 23].
Development of an ARMS-PCR assay targeting the nas 10 nDNA locus
The diagnostic nucleotide positions found between the three Anisakis species were used to develop ARMS-PCR assay experiments. This resulted in the use of 4 pairs of primers organized into 2 primers sets (Table 2). The two sets were specifically designed for these diagnostic nucleotide positions (i.e. 173 and 251) observed at direct sequencing of nas 10 nDNA, in the three species of the A. simplex (s. l.), by using the web-based program, Primers3 plus (https://primer3plus.com/primer3web/primer3web).
Each primer set includes four primers, one pair of primers comprises the forward and reverse outer primers which serve as internal amplification controls; the other pair of primers comprises the forward and reverse inner primers (Table 2). Importantly, in tetra-primer ARMS-PCR, there is not only a 3′ terminus mismatch but also a second deliberate mismatch at position-2 from the 3′ terminal of the inner primers to increase the specificity of the reaction. The two primer sets (Table 2) were used in two PCR reactions planned to work simultaneously, to identify A. pegreffii, A. simplex (s. s.) and A. berlandi. Each PCR was performed in a total volume of 25 μL: 3.5 μL of 10× solution buffer, 1.5 μL containing 10 μM of four mixed dNTPs, 1.5 μL of 50 Mm MgCl2, 1.2 μL of each primer (10 Mm) (Table 2), 0.25 μL of Taq DNA polymerase (5U/μL) (Promega) and 3 μL of total DNA. PCR amplification conditions were as follows: 95 °C for 5 min (initial denaturation) followed by 35 cycles at 95 °C for 30 s (denaturation), 49 °C for 35 s (annealing), 72 °C for 30 s (extension), and a final elongation step at 72 °C for 7 min. The PCR products generated from the outer primers were used as positive controls in the ARMS-PCR system to ensure the normality of PCR reaction (373 bp for primer set-1, 321 bp for primer set-2). The PCR products, obtained from each analyzed Anisakis specimen, by use of two sets of primers (Table 2), were separated by electrophoresis using agarose gel (1.5%) stained with GelRed®; 3 μL of the amplification products were visualized. The distinct banding patterns were detected by the use of ultraviolet transillumination.
Results
Identification of Anisakis simplex (s. l.) specimens
Allozyme analysis of Anisakis (N = 219 specimens) (Table 1) showed that 93 specimens were A simplex (s. s.), 50 A. berlandi, and finally 76 A. pegreffii, according to alleles found at the diagnostic loci between the three sibling species (i.e. Adk-2, Pep C-1, Pep C-2, and Mdh-1) [22]. Similarly, according to the sequences of 629 bp in length of the mtDNA cox2 gene locus [22], 76 specimens were assigned to A. pegreffii, 93 to the species A. simplex (s. s.) and, finally, 50 individuals corresponded to A. berlandi. The sequences obtained in the present study matched, at 100% or 99%, with the sequences previously deposited by us in GenBank for these species. In addition, the identity of these specimens recognized at mtDNA cox2 as belonging to A. pegreffii and A. simplex (s. s.), was confirmed according to the diagnostic positions of the EF1 α-1 nDNA locus [23].
Genotyping of Anisakis spp. by direct sequencing of nas 10 nDNA
A fragment of 451 bp in length of the nas 10 nDNA region was obtained for all of the 219 specimens analyzed here. The sequences revealed the presence of one diagnostic nucleotide site between A. simplex (s. s.) versus A. pegreffii/A. berlandi, and another one between A. berlandi versus A. pegreffii/A. simplex (s. s.): these diagnostic nucleotide positions were 173 and 251, respectively (Figure 1). In particular, position 173 showed the T allele in all the A. simplex (s. s.) individuals (N = 93); while it always showed the C allele in the specimens of A. pegreffii and A. berlandi considered here (Fig. 1). Position 251 showed the G allele in the individuals of A. pegreffii and A. simplex (s. s.), while the allele was always C in all the A. berlandi specimens (N = 50) analyzed here (Fig. 1). All the specimens belonging to the three Anisakis species were found to be homozygous at these nucleotide positions; no heterozygosity between the three species was found in the present study, at these positions. As a consequence, since these nucleotide positions are fixed for alternative nucleotides among the three species, they were considered to have diagnostic value. Sequences of the nas 10 nDNA region were deposited in GenBank under the following accession numbers: MN897674, MN897675, MN897676 for A. pegreffii, MN897671, MN897672, MN897673 for A. simplex (s. s.), and MN897677, MN897678, MN897679 for A. berlandi.
 |
Figure 1 Sequence alignment of the nucleotide sequences of the metallopeptidase 10 (nas 10) of A. simplex (s. s.), A. pegreffii and A. berlandi. Dots indicate identity with the consensus sequence. Arrows indicate the nucleotide diagnostic position. |
Genotyping of Anisakis spp. by ARMS-PCR targeting nas 10 nDNA
The two point mutations diagnostic between the three Anisakis species and the tetra-primer ARMS-PCR analysis enabled us to genotype the individuals belonging to the three species studied here. All the specimens belonging to the three taxa, previously identified by allozymes, mtDNA cox2, and EF1 α-1 nDNA and sequenced at the nas 10 nDNA locus, were directly analyzed by ARMS-PCR at this locus.
One set of designed primers (primer set-1) (Table 2) was able to detect the first point mutation (i.e. 173) making it possible to distinguish A. simplex (s. s.) versus A. pegreffii/A. berlandi; while, the second set of primers detected the point mutation 251, which always identified a C/C genotype in A. berlandi; while it showed a G/G genotype in both A. simplex (s. s.) and A. pegreffii. In particular, in primer set-1 (Table 2) the combined use of the nas 10-reverse outer primer (OUT-R1) and nas 10-forward inner primer (INN-F1), amplifying the T-allele at position 173, generated a specific PCR product of 296 bp in A. simplex (s. s.) (Fig. 2; Table 3). Similarly, the combined use of nas 10-forward outer primer (OUT-F1) and nas 10-reverse inner primer (INN-R1), amplifying the C-allele at position 173, generated a specific PCR fragment of 117 bp in A. pegreffii and A. berlandi specimens (Fig. 2; Table 3). In primer set-2 (Table 2), the combined nas 10-forward outer primer (OUT-F2) and nas 10-reverse inner primer (INN-R2) amplified the G-allele at the diagnostic nucleotide position 251, producing a specific PCR fragment of 148 bp in A. simplex (s. s.) and A. pegreffii (Fig. 2; Table 3). In the same primer set, the nas 10-forward inner primer (INN-F2) and the nas 10-reverse outer primer (OUT-R1) amplified the C-allele (diagnostic position 251) and determined a PCR product of 216 bp in A. berlandi (Fig. 2; Table 3).
 |
Figure 2 Schematic illustration of the two sets of the tetra-primers for the ARMS-PCR design and DNA gel patterns of the different single nucleotide genotyping (on the right). Asterisks indicate the second mismatch of the inner primer. |
Molecular key based on the diagnostic positions.
All the specimens were homozygous for these fixed alternative nucleotide positions; no heterozygosity at these positions was found in the samples of Anisakis spp. tested in this study.
On the basis of the gel electrophoresis patterns (Figure 3) obtained by the combined use of tetra-primer set-1 and tetra-primer set-2, 76 specimens were found to be A. pegreffii, 93 A. simplex (s. s.), and 50 A. berlandi. The findings showed 100% concordance between the genotyping obtained by the ARMS-PCR assay and direct sequence analysis of the same gene locus nas 10 nDNA.
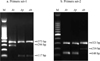 |
Figure 3 Products (genotypes) of tetra-primer ARMS-PCR obtained at the nas 10 locus in the three species of the A. simplex (s. l.) complex. |
Discussion
In parasitic nematodes, peptidase proteins represent around 2% of the genes expressed; they are involved in host-parasite interactions, parasite immune evasion, life cycle transmission, and pathogenesis [19]. In many cases, peptidases are also the main source of parasite antigens, favoring the host immune response; many of them are particularly immunogenic [10, 34]. In particular, metallopeptidases are a group of peptidases that are involved in the invasion of the host tissue by the parasite as they are able to degrade the extracellular matrix. In some cases, it has been suggested that the metallopeptidases conserve their proteolytic activity even after the antibody action exhibited by the host during infection by the parasite [4, 5, 18].
Recent RNA sequence analysis revealed a high percentage of the gene coding for metallopeptidases highly expressed in A. pegreffii third-stage larvae maintained in vitro co-cultured with dendritic cells, at 37 °C (Palomba et al. personal obs.). These enzymes were postulated to be involved in host tissue penetration by Anisakis spp. larvae [11]. The ability of the parasite to migrate to tissue would represent an evolutionary advantage for the larvae of Anisakis spp. during intermediate/paratenic host infection. In turn, this would mean that selective pressure would have conferred an evolutionary advantage to the genes coding for the group of peptidase proteins, allowing the parasite species to successfully invade different fish hosts. As a consequence, a high level of mutations would have occurred in these gene loci over an evolutionary time scale, resulting in an advantageous genotype for the parasite species, in terms of invasion and adaptation to its hosts’ ranges. Interestingly, previous allozyme studies in anisakids have demonstrated that among the scored gene loci, those coding for the peptidase proteins represent a high percentage showing fixed alternative alleles between related anisakid species belonging to the genera Anisakis, Pseudoterranova and Contracaecum [22, 28, 30, 31]. Furthermore, in the case of the three species of the A. simplex (s. l.) complex, the gene locus coding for the enzymatic proteins: Peptidase-C, Peptidase-B, and Leucine-amino-peptidase (i.e. coding loci: Pep-C-1, PepC-2, Pep B, and Lap-1, respectively) showed fixed alternative alleles that are diagnostic in these species. Thus, it seems that these gene loci coding for peptidases are fast evolving loci in anisakids [21], suitable for discriminating between closely related taxa [8].
The possibility of looking directly at the gene sequences coding for these functional proteins, such as peptidases, enables detection of fixed nucleotide positions. These positions are associate with a polymorphic sequence caused by a single nucleotide mutation at a specific locus in the DNA sequence. This type of polymorphism was found through direct sequencing of the gene coding for the metallopeptidase 10 (nas 10 nDNA) presented, which showed alternative single nucleotide positions among the three Anisakis species: i.e. a further diagnostic nuclear gene. A molecular key based on these nucleotide differences is presented in Table 3. No heterozygous genotypes were observed: this is another demonstration that A. pegreffii, A. simplex (s. s.) and A. berlandi are independent gene pools.
The presence of the fixed differences can be considered a Mendelian character because nas 10 nDNA is a nuclear gene locus. This means that these polymorphisms can be also tested for Hardy–Weinberg equilibrium (HWE) and be included in Bayesian clustering multi-genotyping analysis [24, 26], in future multi-locus approaches.
A sufficient sample size (i.e. number of specimens) is required to ensure that the detected fixed differences are maintained even when single specimens from another location are correctly allocated to the correct taxon, i.e. to the three Anisakis species. In the present study, we have validated the existence of the fixed differences in a large number of larval and adult specimens of the three Anisakis species collected from different fish and cetacean hosts.
The present study has also shown that the combined use of two primer sets for ARMS-PCR, established based on the diagnostic polymorphisms at the nas 10 nDNA locus, is suitable for direct and rapid genotype determination of the three Anisakis species. In particular, primer set-1 discriminated the species A. simplex (s. s.) that showed the two fragments at 373 bp and 296 bp, from the species A. pegreffii/A. berlandi, which have the same banding pattern of 373 bp and 117 bp (Table 3, Fig. 3). Then, the primers set-2 allowed the discrimination between A. berlandi with the fragments of 321 bp and 216 bp, versus A. simplex (s. s.)/A. pegreffii (321 bp, 148 bp). Thus, the use of both primers set is mandatory to discriminate between the three Anisakis species (Table 3, Fig. 3).
Tetra-primer ARMS-PCR could be a useful tool for genotyping when direct DNA sequencing could be a time consuming, technically demanding, and costly procedure. For the genetic identification of parasites, such as anisakid nematodes, infecting economically important fish, rapid and cost-effective assays that can be performed with standard PCR instruments are highly desirable. Compared to other genotyping techniques, tetra-primer ARMS-PCR has been reported to be a rapid, reliable, simple and economical assay for SNP genotyping [16, 18, 29, 36]. ARMS can distinguish a single base sequence difference using one-step PCR. This method has recently been developed for SNPs of ITS rDNA for the species identification of A. pegreffii, A. simplex (s. s.) and A. typica occurring in fish from Korean waters [11]. However, it has also recently been suggested that polymorphic nucleotide positions at these loci of the ITS region of rDNA occur, thus confounding, especially in sympatric areas, the correct identification of the nematodes belonging to the species A. pegreffii and A. simplex (s. s.) [24]. Thus, in the present study, a tetra-primer ARMS-PCR based methodology was developed based on the novel nuclear marker, for genotyping A. simplex (s. l.) nematodes. The assay would be also more convenient than the traditional PCR-RFLP since it eliminates the need for incubation with restriction enzymes. This not only avoids any consequent errors and artefacts from such procedures but also reduces the amount of DNA required for the digestion step in PCR-RFLP. No special equipment and only a small amount of standard PCR reagents are needed in tetra-primer ARMS-PCR. The tetra-primer ARMS-PCR protocol described in the present study is the first reported method enabling genotyping of novel SNPs at the nas 10 gene locus. This method is rapid, simple, reliable, easy to perform, and economical, and requires a minimum level of expertise. It can be used for both large- and small-scale genotyping studies. The tetra-primer ARMS-PCR technique could also be developed in the future for other nuclear loci that will show diagnostic nucleotide positions between the three species of the A. simplex (s. l.) complex.
Conclusions
The novel nuclear marker (nas 10 nDNA) and the development of an ARMS-PCR protocol, based on the diagnostic SNPs detected between the three species of Anisakis, proposed in the present study, will allow genotyping of specimens at a further locus when a multilocus-approach is needed. The markers were validated on 219 samples belonging to the three species of Anisakis (i.e. A. pegreffii, A. simplex (s. s.) and A. berlandi). Agreement was observed between the identification and genotyping obtained by the tetra-primer ARMS PCR assay and direct DNA sequencing. The protocols detailed here outline methods that can be used to analyze further genomic DNA of the three sibling species of Anisakis for two diagnostic mutations.
Currently, SNP markers are one of the preferred genotyping approaches, because they are abundant in the genome, genetically stable and amenable to high throughput automated analysis of Anisakis simplex (s. l.) [17]. Diagnostic SNPs in further candidate nuclear genes in the species of A. simplex (s. l.) may be useful in multilocus genotyping protocols required for studying micro-evolutionary aspects. This will help to reach a high number of individuals of the three Anisakis species genotyped at several nuclear loci, including the novel nuclear locus, DNA microsatellites, and other gene loci [3, 24, 26]. This approach will be useful to our further understanding of hybridization and/or introgression phenomena between the species of the A. simplex (s. l.) complex in sympatric areas.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This study was supported by a grant from the “Istituto Pasteur Italia-Fondazione Cenci Bolognetti”. We thank the three anonymous referees for their comments on the manuscript.
References
- Anderson EC, Thompson EA. 2002. A model-based method for identifying species hybrids using multilocus genetic data. Genetics, 160, 1217–1229. [PubMed] [Google Scholar]
- Anderson TJC. 2001. The dangers of using single locus markers in parasite epidemiology: Ascaris as a case study. Trends in Parasitology, 17, 183–188. [CrossRef] [PubMed] [Google Scholar]
- Bello E, Paoletti M, Webb SC, Nascetti G, Mattiucci S. 2020. Cross-species utility of microsatellite loci for the genetic characterization of Anisakis berlandi (Nematoda: Anisakidae). Parasite, 27, 9. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Benitez R, Kasny M, Adroher FJ. 2013. Peptides in parasitic nematodes, in . Parasites: Ecology, Diseases and Management, Erzinger GS, Editor. Nova Science Publishers Inc.: United Kingdom, 61–102. [Google Scholar]
- Bruschi F, Pinto B. 2017. Matrix metalloproteinases in parasitic infection, in Pathophysiological aspects of proteases. Chakraborti S, Dhalla NS, Editors. Springer: Canada. p. 321–352. [CrossRef] [Google Scholar]
- Cipriani P, Acerra V, Bellisario B, Sbaraglia GL, Cheleschi R, Nascetti G, Mattiucci S. 2016. Larval migration of the zoonotic parasite Anisakis pegreffii (Nematoda: Anisakidae) in European anchovy, Engraulis encrasicolus: Implications to seafood safety. Food Control, 59, 148–157. [Google Scholar]
- Criscione CD, Anderson JD, Raby K, Sudimack D, Subedi J, Rai DR, Upadhayay RP, Jha B, Williams-Blangero S, Anderson T. 2007. Microsatellite markers for the human nematode parasite Ascaris lumbricoides: development and assessment of utility. Journal of Parasitology, 93, 704–708. [CrossRef] [Google Scholar]
- Ferguson A. 1980. Biochemical systematics and evolution. John Wiley and Sons: New York. [Google Scholar]
- Guardone L, Armani A, Nucera D, Costanzo F, Mattiucci S, Bruschi F. 2018. Human anisakiasis in Italy: a retrospective epidemiological study over two decades. Parasite, 25, 41. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Hotez PJ, Pritchard DI. 1995. Hookworm infection. Scientific American, 272, 68–74. [CrossRef] [PubMed] [Google Scholar]
- Kim H, Baek KW, Park MK, Jeon KJ, Ko EJ, Cha HJ, Ock MS. 2019. Establishment and validation of ARMS (amplification-refractory mutation system) for identification of Anisakis species collected from Korean water. Gene, 691, 125–131. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [CrossRef] [PubMed] [Google Scholar]
- Levsen A, Cipriani P, Mattiucci S, Gay M, Lee CH, MacKenzie K, Pierce GJ, Svanevik CS, Hojgaard DP, Nascetti G, Gonzalez AF, Pascual S. 2018. Anisakis species composition and infection characteristics in Atlantic mackerel, Scomber scombrus, from major European fishing grounds – reflecting changing fish host distribution and migration pattern. Fisheries Research, 202, 112–121. [Google Scholar]
- Levsen A, Svanevik CS, Cipriani P, Mattiucci S, Gay M, Hastiee LC, Pierce GJ, Bušelić I, Mladineo I, Karl O, Ostermeyer U, Buchmann K, Højgaard DP, González AF, Pascual S. 2018. A survey of zoonotic nematodes of commercial key fish species from major European fishing grounds – introducing the FP7 PARASITE exposure assessment study. Fisheries Research, 202, 4–21. [Google Scholar]
- Lim H, Jung BK, Cho J, Yooyen T, Shin EH, Chai JY. 2015. Molecular diagnosis of cause of anisakiasis in humans. South Korea. Emerging Infectious Diseases, 21, 342. [CrossRef] [PubMed] [Google Scholar]
- Little S. 2001. Amplification-refractory mutation system (ARMS) analysis of point mutations. Current Protocol in Human Genetics, 9, 1–8. [Google Scholar]
- Lopienska-Biernat E, Paukszto L, Jastrzebski JP, Myszczynski K, Polak I, Stryinski R. 2019. Genome-wide analysis of Anisakis simplex sensu latu: the role of carbohydrate metabolism genes in the parasite’s development. International Journal for Parasitology, 49, 933–943. [CrossRef] [PubMed] [Google Scholar]
- Malagón D, Benítez R, Adroher FJ, Díaz-López M. 2011. Proteolytic activity in Hysterothylacium aduncum (Nematoda: Anisakidae), a fish gastrointestinal parasite of worldwide distribution. Veterinary Parasitology, 183, 95–102. [CrossRef] [PubMed] [Google Scholar]
- Malagón D, Benitez R, Kasny M, Adroher FJ. 2013. Peptidases in parasitic nematodes. A review, in Parasites: Ecology, diseases and management. Erzinger GS, Editor. United Kingdom. p. 61–102. [Google Scholar]
- Mattiucci S, Nascetti G, Cianchi R, Paggi L, Arduino P, Margolis L, Brattey J, Webb SC, D’Amelio S, Orecchia P, Bullini L. 1997. Genetic and ecological data on the Anisakis simplex complex with evidence for a new species (Nematoda, Ascaridoidea, Anisakidae). Journal of Parasitology, 83, 401–416. [CrossRef] [Google Scholar]
- Mattiucci S, Nascetti G. 2008. Advances and trends in the molecular systematics of anisakids nematodes, with implications for their evolutionary ecology and host-parasite co-evolutionary processes. Advances in Parasitology, 66, 47–148. [CrossRef] [PubMed] [Google Scholar]
- Mattiucci S, Paoletti M, Webb SC. 2009. Anisakis nascettii n. sp. (Nematoda: Anisakidae) from beaked whales of the southern hemisphere: morphological description, genetic relationships between congeners and ecological data. Systematic Parasitology, 74, 199–217. [CrossRef] [PubMed] [Google Scholar]
- Mattiucci S, Cipriani P, Webb SC, Paoletti M, Marcer F, Bellisario B, Gibson DI, Nascetti G. 2014. Genetic and morphological approaches distinguishing the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi n. sp. for A. simplex sp. C (Nematoda: Anisakidae). Journal of Parasitology, 15, 12–15. [Google Scholar]
- Mattiucci S, Acerra V, Paoletti M, Cipriani P, Levsen A, Webb SC, Canestrelli D, Nascetti G. 2016. No more time to stay “single” in the detection of Anisakis pegreffii, A. simplex (s. s.) and hybridization events between them: a multi-marker nuclear genotyping approach. Parasitology, 143, 998–1011. [CrossRef] [PubMed] [Google Scholar]
- Mattiucci S, Cipriani P, Levsen A, Paoletti M, Nascetti G. 2018. Molecular epidemiology of Anisakis and anisakiasis: an ecological and evolutionary road map. Advances in Parasitology, 99, 93–263. [CrossRef] [PubMed] [Google Scholar]
- Mattiucci S, Bello E, Paoletti M, Webb SC, Timi JT, Levsen A, Cipriani P, Nascetti G. 2019. Novel polymorphic microsatellite loci in Anisakis pegreffii and A. simplex (s. s.) (Nematoda: Anisakidae): implications for species recognition and population genetic analysis. Parasitology (11), 1–17. [Google Scholar]
- Mladineo I, Trumbić Ž, Radonić I, Vrbatović A, Hrabar J, Bušelić I. 2017. Anisakis simplex complex: ecological significance of recombinant genotypes in an allopatric area of the Adriatic Sea inferred by genome-derived simple sequence repeats. International Journal for Parasitology, 47, 215–223. [CrossRef] [PubMed] [Google Scholar]
- Nascetti G, Cianchi R, Mattiucci S, D’Amelio S, Orecchia P, Paggi L, Brattey J, Berland B, Smith JW, Bullini L. 1993. Three sibling species within Contracaecum osculatum (Nematoda, Ascaridida, Ascaridoidea) from the Atlantic Arctic-Boreal region: Reproductive isolation and host preferences. International Journal of Parasitology, 23, 105–120. [CrossRef] [Google Scholar]
- Okayama N, Fujimura K, Nakamura J, Suehiro Y, Hamanaka Y, Hinoda Y. 2004. Evaluation of a new efficient procedure for single-nucleotide polymorphism genotyping: tetra-primer amplification refractory mutation system-polymerase chain reaction. Journal of Clinical Chemistry and Laboratory Medincine, 42, 13–16. [Google Scholar]
- Orecchia P, Mattiucci S, D’Amelio S, Paggi L, Plotz J, Cianchi R, Nascetti G, Arduino P, Bullini L. 1994. Two new members in the Contracaecum osculatum complex (Nematoda, Ascaridoidea) from the Antarctic. International Journal for Parasitology, 24, 367–377. [CrossRef] [PubMed] [Google Scholar]
- Paggi L, Nascetti G, Cianchi R, Orecchia P, Mattiucci S, D’Amelio S, Berland B, Brattey J, Smith JW, Bullini L. 1991. Genetic evidence for three species within Pseudoterranova decipiens (Nematoda, Ascaridida, Ascaridoidea) in the North Atlantic and Norwegian and Barents Seas. International Journal of Parasitology, 21, 195–212. [CrossRef] [Google Scholar]
- Patel YM, Newton CR, Markham AF, Fleming KA, Wainscoat JS. 1991. Direct haplotype determination by double ARMS: specificity, sensitivity and genetic applications. Nucleic Acids Research, 19, 3561–3567. [CrossRef] [PubMed] [Google Scholar]
- Pontes T, D’Amelio S, Costa G, Paggi L. 2005. Molecular characterization of larval anisakid nematodes from marine fishes of Madeira by a PCR-based approach, with evidence for a new species. Journal of Parasitology, 91, 1430–1434. [CrossRef] [Google Scholar]
- Todorova VK, Stoyanov DI. 2000. Partial characterization of serine proteinases secreted by adult Trichinella spiralis. Parasitology Research, 86, 684–687. [CrossRef] [PubMed] [Google Scholar]
- Valentini A, Mattiucci S, Bondanelli P, Webb SC, Mignucci-Giannone A, Colom-Llavina MM, Nascetti G. 2006. Genetic relationships among Anisakis species (Nematoda: Anisakidae) inferred from mitochondrial cox2 sequences, and comparison with allozyme data. Journal of Parasitology, 92, 156–166. [CrossRef] [Google Scholar]
- Ye S, Dhillon S, Ke X, Collins AR, Day INM. 2001. An efficient procedure for genotyping single nucleotide polymorphism. Nucleic Acids Research, 29, 17. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Palomba M, Paoletti M, Webb SC, Nascetti G & Mattiucci S. 2020. A novel nuclear marker and development of an ARMS-PCR assay targeting the metallopeptidase 10 (nas 10) locus to identify the species of the Anisakis simplex (s. l.) complex (Nematoda, Anisakidae). Parasite 27, 39.
All Tables
Sampling area, host species and life-history stage (A: adult; L4: 4th stage larvae; L3: 3rd stage larvae) of the specimens of A. simplex (s. s.), A. pegreffii and A. berlandi analyzed.
Primer sequences targeting the metallopeptidase 10 (nas 10) locus, product size, and annealing temperature (T a/°C). In bold, a deliberate second mismatch.
All Figures
 |
Figure 1 Sequence alignment of the nucleotide sequences of the metallopeptidase 10 (nas 10) of A. simplex (s. s.), A. pegreffii and A. berlandi. Dots indicate identity with the consensus sequence. Arrows indicate the nucleotide diagnostic position. |
| In the text | |
 |
Figure 2 Schematic illustration of the two sets of the tetra-primers for the ARMS-PCR design and DNA gel patterns of the different single nucleotide genotyping (on the right). Asterisks indicate the second mismatch of the inner primer. |
| In the text | |
 |
Figure 3 Products (genotypes) of tetra-primer ARMS-PCR obtained at the nas 10 locus in the three species of the A. simplex (s. l.) complex. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


