| Issue |
Parasite
Volume 25, 2018
|
|
|---|---|---|
| Article Number | 29 | |
| Number of page(s) | 10 | |
| DOI | https://doi.org/10.1051/parasite/2018031 | |
| Published online | 28 May 2018 | |
urn:lsid:zoobank.org:pub:965C0D25-C3D4-4A55-99F2-0EB7726CDA8D
Research Article
Balantidium grimi n. sp. (Ciliophora, Litostomatea), a new species inhabiting the rectum of the frog Quasipaa spinosa from Lishui, China
Balantidium grimi n. sp. (Ciliophora, Litostomatea), nouvelle espèce habitant le rectum de la grenouille Quasipaa spinosa de Lishui, Chine
1
Key Laboratory of Aquaculture Disease Control, Ministry of Agriculture, and State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences,
430072
Wuhan, PR China
2
Hubei Key Laboratory of Animal Nutrition and Feed Science, Wuhan Polytechnic University,
430023
Wuhan, PR China
3
University of the Chinese Academy of Sciences,
100049
Beijing, PR China
4
Animal Husbandry and Aquaculture Station, Agriculture Forestry Animal Husbandry and Aquaculture Bureau of Guye District of Tangshan City,
063100
Tangshan, PR China
* Corresponding author: liming@ihb.ac.cn.
Received:
27
January
2018
Accepted:
1
May
2018
Balantidium grimi n. sp. is described from the rectum of the frog Quasipaa spinosa (Amphibia, Dicroglossidae) from Lishui, Zhejiang Province, China. The new species is described by both light microscopy (LM) and scanning electron microscopy (SEM), and a molecular phylogenetic analysis is also presented. This species has unique morphological features in that the body shape is somewhat flattened and the vestibulum is “V”-shaped, occupying nearly 3/8 to 4/7 of the body length. Only one contractile vacuole, situated at the posterior body, was observed. The phylogenetic analysis based on SSU-rDNA indicates that B. grimi groups together with B. duodeni and B. entozoon. In addition, the genus Balantidium is clearly polyphyletic.
Résumé
Balantidium grimi n. sp. est décrit à partir du rectum de la grenouille Quasipaa spinosa (Amphibia, Dicroglossidae) de Lishui, province de Zhejiang, Chine. La nouvelle espèce est décrite à la fois par microscopie photonique et électronique à balayage, et une analyse phylogénétique moléculaire est également présentée. Cette espèce possède des caractéristiques morphologiques uniques en ce que la forme du corps est quelque peu aplatie et que le vestibule est en forme de « V » et occupe près de 3/8 à 4/7 de la longueur du corps. Une seule vacuole contractile, située au niveau du corps postérieur, a été observée. L’analyse phylogénétique basée sur la petite sous-unité de l’ADNr indique que B. grimi forme un groupe avec B. duodeni et B. entozoon. De plus, le genre Balantidium est clairement polyphylétique.
Key words: Balantidium grimi / ciliate / new species / Quasipaa spinosa / China
© W. Zhao et al., published by EDP Sciences, 2018
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
1 Introduction
The genus Balantidium Claparède & Lachmann, 1858 consists of many species inhabiting the digestive tract in a wide number of hosts from both invertebrate and vertebrate animals as endocommensals. They are generally considered harmless, but factors depressing the resistance of the host enable them to invade the mucosa and cause ulceration. The representatives of Balantidium have some common morphological features: cell body sacciform or slightly elongated in shape, and completely covered with cilia forming dense longitudinal rows [21]. To our knowledge, 31 amphibian balantidial species have been reported so far (lists in Li et al. [20]).
To date, 27 valid species have been reported in anuran amphibians, including B. amygdalli Bhatia & Gulati, 1927 [3], B. aurangabadensis Shete & Krishnamurthy, 1984 [34], B. bicavata Bhatia & Gulati, 1927 [3], B. claperedei Mahoon & Khan, 1986 [22], B. corlissi Shete & Krishnamurthy, 1984 [34], B. cyanophlycti Shete & Krishnamurthy, 1984 [34], B. duodeni Stein, 1867 [36], B. elongatum Stein, 1867 [36], B. entozoon Ehrenberg, 1838 [9], B. falciformis Walker, 1909 [40], B. ganapatii Shete & Krishnamurthy, 1984 [34], B. giganteum Bezzenberger, 1904 [2], B. gracile Bezzenberger, 1904 [2], B. helenae Bezzenberger, 1904 [2], B. honghuensis Li et al., 2013 [18], B. kirbyi Rodriguez, 1939 [31], B. megastomae Shete & Krishnamurthy, 1984 [34], B. mininucleatum Shete & Krishnamurthy, 1984 [34], B. ranae Shete & Krishnamurthy, 1984 [34], B. ranarum Ghosh, 1921 [10], B. rotundum Bezzenberger, 1904 [2], B. sinensis Nie, 1935 [24], B. singaporensis Khan & Ip, 1986 [16], B. sushilii Ray, 1932 [30], B. tigrinae Shete & Krishnamurthy, 1984 [34], B. vanensis Senler & Yildiz, 2000 [33] and B. xenopi Puytorac & Grain, 1965 [28]. Five other balantidial species were found in urodele amphibians, including B. amblystomatis Jírovec, 1930 [15], B. andianusis Li et al., 2008 [20], B. elongatum Stein, 1867 [36], B. rayi Pal & Dasgupta, 1978 [25] and B. tylototritonis Pal & Dasgupta, 1978 [25]. Among the aforementioned species, 3 balantidial species inhabiting amphibians were first discovered and named in China. B. andianusis was reported in the Chinese giant salamander, Andrias davidianus [20]; B. sinensis was described from 2 species of anuran amphibians and 1 urodele amphibian, R. nigromaculata, R. plancyi [24] and A. davidianus [20], respectively, and B. honghuensis was found in R. nigromaculata [18].
Although many amphibian Balantidium species have been reported, few molecular data are available at present (only two species B. entozoon and B. duodeni have corresponding SSU-rDNA sequences in NCBI). Even less is known about phylogenetic relationships between different balantidial groups inhabiting different hosts (such as fishes, amphibians, mammals, etc.).
In the present study, a new Balantidium species inhabiting Quasipaa spinosa is described based on detailed light and scanning electron microscopy observation. This is also the first record of Balantidium species in the digestive tract of Quasipaa spinosa. Phylogenetic analysis based on SSU-rDNA was also carried out to reveal the relationships among Balantidium species as well as different clades of Trichostomatia.
2 Materials and methods
2.1 Specimen collection and identification
The frogs used for this study were captured from Lishui City (27°25′–28°57′ N, 118°41′–120°26′ E), Zhejiang Province, southeast China in August, 2017. We obtained permits allowing us to capture and sacrifice these specimens. The frogs were transported alive to the laboratory, then all frog samples were anesthetized and dissected as soon as possible, the luminal contents of recta, intestines and duodena were collected respectively into different Petri dishes, and examined with the help of a stereomicroscope (Leica S8AP0, Germany). The ciliates were collected with Pasteur micropipettes and washed twice in 0.65% NaCl solution.
2.2 Light microscopy
Some specimens were fixed in 5% formalin for 10 min and soaked for about 30 min in 10% glycerin alcohol in a concave slide; the remaining specimens were fixed in Bouin’s fluids and stained with a Protargol method [11]. Specimens were observed, measured and photographed using a microscope (Olympus BX53, Japan). All measurements are in micrometers.
2.3 Scanning electron microscopy
The fully washed specimens were fixed in 2.5% glutaraldehyde in 0.2 M PBS (pH 7.4) on a clean glass slide (1 cm × 1 cm), which was previously treated with 0.1% poly-L-Lysine and dried completely in the air at room temperature. After being washed with PBS 3 times, they were post-fixed in 1% osmium tetroxide at 4°C for 1 h, followed by serial dehydration in acetone and critical point drying using the HCP-2 critical point dryer (Hitachi Science Systems, Japan). Subsequently, the glass slide was mounted on an aluminum-stub using a double-sided adhesive tape and sputter-coated with a thin layer of gold in IB-3 ion coater (Eiko Engineering, Japan), before observation and photography using a Quanta 200 SEM (FEI, Netherlands).
2.4 Extraction of genomic DNA and PCR amplification
About 50 individuals were harvested, suspended in lysis buffer (10 mM Tris-HCl, pH 8.0; 1 M EDTA, pH 8.0; 0.5 % sodium dodecyl sulfate; 60 µg/mL proteinase K), and incubated at 55°C for 12–20 h. DNA was extracted using a standard phenol/chloroform method, precipitated with ethanol, and resuspended in TE buffer. Polymerase chain reaction (PCR) amplifications were carried out using forward primer (5’-AACCTGGTTGATCCTGCCAGT-3’) and reverse primer (5’-TGATCCTTCTGCAGGTTCACCTAC-3’) [23]. The following cycling conditions included 5 min initial denaturation at 94°C; 35 cycles of 30s at 95°C, 1 min at 56-60 °C, and 1-2 min at 72°C; with a final extension of 10 min at 72°C. The PCR products were isolated using 1% agarose gel electrophoresis and purified using the Agarose Gel DNA Purification Kit (TaKaRa Biotechnology, Dalian, Japan). The amplified fragment was cloned into a pMD®18-T vector (TaKaRa Biotechnology, Dalian) and sequenced in both directions using M13 forward and reverse primers on an ABI PRISM® 3730 DNA Sequencer (Applied Biosystems, USA). The SSU rRNA gene sequence of B. grimi was deposited in GenBank with accession number MG837094.
2.5 Phylogenetic analysis
Besides the SSU-rDNA sequence of B. grimi that we obtained in this study, other litostomatean sequences were retrieved from the GenBank/EMBL databases (Table 1). The sequence of Nyctotheroides deslierresae was used as the outgroup. The secondary structure-based SSU-rRNA sequence alignment of Litostomatea downloaded from the SILVA ribosomal RNA gene database project (https://www.arb-silva.de/) [29] was used as the “seed” alignment to build a profile Hidden Markov Model (HMM) using HMMER Package, version 3.1. Then the HMM profile obtained was used to create an alignment of the 40 sequences using Hmmalign within the package. The masked regions that could not be aligned unambiguously were removed from the initial alignment using MEGA 6.0 [39]. A GTR+I+G model was selected as the best model by the program jModelTest 2.1.10 [8] based on the AIC criterion, which was used for both Maximum Likelihood (ML) and Bayesian (BI) inference analysis. An ML tree was constructed with the RaxML program [35]. The reliability of internal branches was assessed using the non-parametric bootstrap method with 1,000 pseudoreplicates. A Bayesian analysis performed with MrBayes v3.2.6 [32] was run for 1,000,000 generations sampling every 1,000 generations. All trees below the observed stationary level were discarded as a burn-in of 25% of the generations.
List of sequences from GenBank/EMBL databases used for phylogenetic analysis.
3 Results
Ninety-eight individuals of Q. spinosa were examined in the present study and 34 were found to be infected with Balantidium grimi (prevalence, 34.7%). These specimens were found mainly in the recta of frogs.
Balantidium grimi n. sp.
urn:lsid:zoobank.org:act:84E00073-0D0C-4166-8D83-20BFCC43480E
Type host: Quasipaa spinosa David, 1875.
Prevalence: 34.7% (34 of 98) of Q. spinosa were infected.
Type locality: Lishui City (27°25′–28°57′N, 118°41′–120°26′E), Zhejiang Province, China.
Infection site: Rectum.
Type material: Holotype catalogued under No. IHB2017W005, paratype catalogued under No. IHB2017W006 with protargol stained and the rest of ciliates preserved in 100% alcohol (Nos. LS001-002), 2.5% glutaraldehyde (No. LS003) and Bouin’s fluids (Nos. LS004-LS006) have been deposited in Key laboratory of Aquaculture Disease Control, Ministry of Agriculture, Institute of Hydrobiology, Chinese Academy of Sciences, China.
Etymology: The new species was designated Balantidium grimi n. sp. in honor of the great contributions of Prof. J. Norman Grim to parasitic and symbiotic ciliates.
3.1 Morphology under light microscope
Organism long-oval in shape (Figures 1A, C and 2), measuring 79.6-121.5µm ( = 96.5 µm; n = 30) in length and 43.6-83.6 µm (
= 96.5 µm; n = 30) in length and 43.6-83.6 µm (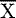 = 57.8 µm) in width. Body partially flattened and thickly ciliated (Figures 1A, C and 2). The number of body kineties ranged from 93 to 125, oriented mostly parallel to the cell’s long axis. Of these, 41 to 59 were dorsal and 52 to 67 were ventral. Vestibulum “V”-shaped, 32.6-53.9 µm (
= 57.8 µm) in width. Body partially flattened and thickly ciliated (Figures 1A, C and 2). The number of body kineties ranged from 93 to 125, oriented mostly parallel to the cell’s long axis. Of these, 41 to 59 were dorsal and 52 to 67 were ventral. Vestibulum “V”-shaped, 32.6-53.9 µm ( = 43.43 µm, n = 30) in length, accounted for 3/8 to 4/7 of the body length (Figures 1B, D, E and 2), and 3.9-5.9 µm (
= 43.43 µm, n = 30) in length, accounted for 3/8 to 4/7 of the body length (Figures 1B, D, E and 2), and 3.9-5.9 µm ( = 4.7 µm, n = 30) in width. Macronucleus oval and lay obliquely almost near the middle of body (Figures 1C, E, F and 2), 20.0-29.2 µm (
= 4.7 µm, n = 30) in width. Macronucleus oval and lay obliquely almost near the middle of body (Figures 1C, E, F and 2), 20.0-29.2 µm ( = 24.1 µm, n = 30) in length and 12.4-19.3 µm (
= 24.1 µm, n = 30) in length and 12.4-19.3 µm ( = 16.0 µm, n = 30) in width. Micronucleus spherical or somewhat oval near the macronucleus (Figures 1C, E, F and 2), measuring approximately 2.2-2.9 µm (
= 16.0 µm, n = 30) in width. Micronucleus spherical or somewhat oval near the macronucleus (Figures 1C, E, F and 2), measuring approximately 2.2-2.9 µm ( = 2.5 µm, n = 13) in diameter. A distinct contractile vacuole situated at the posterior region of the body with 12.4-15.4 µm (
= 2.5 µm, n = 13) in diameter. A distinct contractile vacuole situated at the posterior region of the body with 12.4-15.4 µm ( = 13.7 µm, n = 8) in diameter (Figures 1A and 2). A cytoproct present at the posterior end of the body (Figures 1A and 2). Detailed morphometric parameters are presented in Table 2.
= 13.7 µm, n = 8) in diameter (Figures 1A and 2). A cytoproct present at the posterior end of the body (Figures 1A and 2). Detailed morphometric parameters are presented in Table 2.
 |
Figure 1 LM images of B. grimi. A. Specimens fixed in formalin (5%) and soaked in glycerine-alcohol (10%), showing the oval body shape, vestibulum (vb) and macronucleus (ma), a round contractile vacuole (cv) in the posterior and a cytoproct (cp) at the end of the body. Scale bar = 10 µm. B. Specimens fixed in formalin (5%) and soaked in glycerine-alcohol (10%), showing the long vestibulum (vb) surrounded by cilia. Scale bar = 10 µm. C-F. are protargol stained: C. showing the body shape, macronucleus (ma) and micronucleus (mi). Scale bar = 10 µm. D. showing the vestibulum and somatic kineties. Scale bar = 10 µm. E. showing the vestibulum (vb) and the oval macronucleus (ma) with a spherical micronucleus (mi) embedded in the middle. Scale bar = 10 µm. F. showing the relative position of macronucleus (ma) and micronucleus (mi). Scale bar = 5 µm. |
 |
Figure 2 Schematic drawing of B. grimi, showing the general form and structures from the ventral-left view: vestibulum (vb), food particles (fp), macronucleus (ma), micronucleus (mi), contractile vacuole (cv) and cytoproct (cp). Scale bar = 10 µm. |
Morphometric light microscopic parameters of B. grimi.
3.2 Morphology under scanning electron microscope
B. grimi is thickly ciliated, but with uniform arrangement on the cell surface (Figures 3A, B). Regular beat patterns of cilia that look like “waves” make the cell move smoothly (Figure 3A). The “waves” and ridges formed an angle ranging from 0° (at the posterior) to 60° (at the anterior) (Figures 3A, C, D). Numerous cortical grooves arranged alternately with cortical ridges, which are parallel to the longitudinal axis of the body (Figure 3D). The cilia originate within grooves and are quite close together; those in Figure 3D are about 0.62 µm apart.
 |
Figure 3 SEM images of B. grimi. A. Overview of the ventral-left side (oral side), showing the general form, vestibulum (arrow) and uniformly arranged cilia. Scale bar = 10 µm. B. Overview of the right side, showing the body surface is partially flattened and thickly ciliated. Scale bar = 10 µm. C. Ventral-left view of the “V”-shaped vestibulum (arrow). Scale bar = 5 µm. D. The left anterior area of ciliate, showing the vestibulum (vb), an interkinetal ridge (rd), the groove (gr) and the cilia (cl) extending from grooves and are close to one another. Scale bar = 5 µm. E. Selected enlargement of Figure 3D, showing a ridge (rd) between cilia. Scale bar = 2 µm. |
3.3 Phylogenetic analysis
The sequenced SSU-rRNA gene of B. grimi is 1,640 bases in length and the guanine-cytosine (GC) content is 42.26%. The topologies of our phylogenetic trees generated using MrBayes and PhyML algorithms are totally accordant (Figure 4). Species of the family Balantidiidae are separated into three clades. B. grimi grouped together with B. duodeni and the type species of the genus, B. entozoon, and form the first clade whose hosts are anuran amphibians (100% ML, 1.00 BI). B. polyvacuolum and B. ctenopharyngodoni form the second balantidial clade inhabiting fish hosts. The third group consisted of two isolates of B. coli, which were reported from many mammalian hosts, including pigs and humans.
 |
Figure 4 Phylogenetic relationships of the SSU-rRNA sequences of B. grimi marked in bold and other Trichostomatia species showing the position of B. grimi inferred by maximum likelihood method and Bayesian algorithm. The trees were rooted using the sequence of Nyctotheroides deslierresae as the outgroup taxa. Numbers at nodes indicate bootstrap percentage and posterior probability, respectively. The sequences corresponding to species of the genus Balantidium are shadowed. |
4 Discussion
A new Balantidium species inhabiting Chinese anuran amphibians Quasipaa spinosa is recorded herein. To our knowledge, this is the first report of Balantidium species in Q. spinosa.
B. grimi is quite unique considering its remarkably flattened body and conspicuous slit-shaped vestibulum, which can distinguish it from other Balantidium species [7,12,21]. B. grimi resembles B.entozoon, B. duodeni, B. helenae and B. sinensis in some aspects. For example, B. grimi shares a similar Lv/Lb value with B. duodeni [7]. But in terms of body forms and dimensions, these two balantidial species could easily be discriminated from each other. As to the shape and dimension of the macronucleus, as well as the position of the contractile vacuole, B. grimi somewhat resembles B. helenae [33], but the latter species possesses a remarkable “knob” at the posterior end. Comparisons were also made between B. grimi and B. sinensis inhabiting the Chinese giant salamander Andrias davidianus [20] as well as B. entozoon, the type species of the genus Balantidium [12]. Detailed comparisons of morphometric parameters among corresponding Balantidium species are presented in Table 3.
According to the molecular phylogenetic analysis, the order Macropodiniida ciliates is closely related to fish balantidial species [14,19]. The affinity implies that macropodiniids may have been the result of separate invasions of terrestrial hosts by ciliates initially associated with aquatic hosts [19]. Macropodiniids, previously called “Australian clade”, possess similar oral cavities to some vestibuliferids that are bordered by somatic kineties and analogous ultrastructure to the Isotrichidae [5,21,37,38]. Moreover, the strong molecular support of Macropodiniida assemblage as a sister clade to the Balantidiidae (fish balantidia) also gives us an indication that Macropodiniida ought to be incorporated into the order Vestibuliferida, which also coincides with the viewpoint of former studies [5,14,19].
Our results show that the genus Balantidium is clearly polyphyletic and all Balantidium species are separated into three distinct clades, according to host specificity: fish balantidia (B. ctenopharyngodoni and B. polyvacuolum), amphibian balantidia (B. grimi, B. entozoon and B. duodeni), and balantidia from warm-blooded vertebrates (Balantioides coli) [7]. Pomajbíková et al. [26] has proposed a new genus Neobalantidium for the third group. However, it was recently suggested to reinstate the genus Balantioides as this taxon has been named for a long time [7]. Here, we accepted the generic name Balantioides to describe this group. As to the amphibian balantidia, our new species clustered with the other two species, B. entozoon and B. duodeni with maximum molecular supports. On this point, our results are consistent with those of Chistyakova et al. [7], but differ from those of Li et al. [19]. We suspect that the key reason for this disagreement is the quantity of introduced species used for phylogenetic analysis. The greater the number of related species studied, the greater the accuracy of the resulting phylogeny. Thus, more molecular information on Balantidium species from fishes and amphibians as well as reptiles is needed to clarify their phylogenetic relationships.
Comparison of body length (Lb), vestibulum length (Lv) and the ratio of vestibulum length and body length (Lv/Lb) between B. grimi and four Balantidium species.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
Financial support for this study was provided by the National Natural Science Foundation of China (No. 31772429, 31471978), the Youth Innovation Promotion Association CAS (No. Y82Z01), and the Earmarked Fund for China Agriculture Research System (No. CARS-45-15).
References
- Affa’a FL, Hickey DA, Strüder-Kypke M, Lynn DH. 2004. Phylogenetic position of species in the genera Anoplophrya, Plagiotoma, and Nyctotheroides (Phylum Ciliophora), endosymbiotic ciliates of annelids and anurans. Journal of Eukaryotic Microbiology, 51(3), 301-306. [Google Scholar]
- Bezzenberger E. 1904. Über Infusorien aus asiatischen Anuren. Archiv für Protistenkunde, 3, 138-174. [Google Scholar]
- Bhatia BL, Gulati AN. 1927. On some parasitic ciliates from Indian frogs, toads, earthworms and cockroaches. Archiv für Protistenkunde, 57, 85-120. [Google Scholar]
- Cameron SL, Adlard RD, O’Donoghue PJ. 2001. Evidence for an independent radiation of endosymbiotic litostome ciliates within Australian marsupial herbivores. Molecular Phylogenetics and Evolution, 20(2), 302-310. [CrossRef] [PubMed] [Google Scholar]
- Cameron SL, O’Donoghue PJ. 2004. Phylogeny and biogeography of the “Australian” trichostomes (Ciliophora: Litostomata). Protist, 155(2), 215-235. [CrossRef] [PubMed] [Google Scholar]
- Cameron SL, Wright A-DG, O’Donoghue PJ. 2003. An expanded phylogeny of the Entodiniomorphida (Ciliophora: Litostomatea). Acta Protozoologica, 42(1), 1–6. [Google Scholar]
- Chistyakova LV, Kostygov AY, Kornilova OA, Yurchenko V. 2014. Reisolation and redescription of Balantidium duodeni Stein, 1867 (Litostomatea, Trichostomatia). Parasitology Research, 113(11), 4207-4215. [CrossRef] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9(8), 772. [CrossRef] [Google Scholar]
- Ehrenberg CG. 1838. Die infusionsthierchen als vollkommene organismen. Leipzig: Leopold Voss. p. 612. [Google Scholar]
- Ghosh EN. 1921. Infusoria from the environment of Calcutta. I. Bulletin of the Carmichael Medical College, Calcutta, 2, 6-17. [Google Scholar]
- Grim JN. 1988. A somatic kinetid study of the pycnotrichid ciliate Vestibulongum corlissi NG, N. Sp. (Class: Litostomatea), symbiont in the intestines of the surgeonfish, Acanthurus xanthopterus. Journal of Eukaryotic Microbiology, 35(2), 227-230. [Google Scholar]
- Grim JN, Buonanno F. 2009. A re-description of the ciliate genus and type species, Balantidium entozoon. European Journal of Protistology, 45(3), 174-182. [CrossRef] [PubMed] [Google Scholar]
- Irbis C, Garriga R, Kabasawa A, Ushida K. 2008. Phylogenetic analysis of Troglodytella abrassarti isolated from Chimpanzees (Pan troglodytes verus) in the wild and in captivity. Journal of General and Applied Microbiology, 54(6), 409–413. [CrossRef] [Google Scholar]
- Ito A, Ishihara M, Imai S. 2014. Bozasella gracilis n. sp. (Ciliophora, Entodiniomorphida) from Asian elephant and phylogenetic analysis of entodiniomorphids and vestibuliferids. European Journal of Protistology, 50(2), 134-152. [CrossRef] [PubMed] [Google Scholar]
- Jírovec O. 1930. Über ein neues Balantidium aus dem Darmtraktus von Amblystoma tigrinum. Zeitschrift für Parasitenkunde, 3(1), 17-21. [CrossRef] [Google Scholar]
- Khan MM, Ip YK. 1986. Parasites of toads from Singapore, with a description of Balantidium singaporensis sp. n. (Ciliophora: Balantidiidae). Zoological Science, 3(3), 543-546. [Google Scholar]
- Leipe DD, Bernhard D, Schlegel M, Sogin ML. 1994. Evolution of 16S-like ribosomal RNA genes in the ciliophoran taxa Litostomatea and Phyllopharyngea. European Journal of Protistology, 30(3), 354-361. [CrossRef] [Google Scholar]
- Li M, Li W, Zhang L, Wang C. 2013. Balantidium honghuensis n. sp. (Ciliophora: Trichostomatidae) from the rectum of Rana nigromaculata and R. limnocharis from Honghu Lake, China. Korean Journal of Parasitology, 51(4), 427-431. [CrossRef] [Google Scholar]
- Li M, Ponce-Gordo F, Grim JN, Wang C, Nilsen F. 2014. New insights into the molecular phylogeny of Balantidium (Ciliophora, Vetibuliferida) based on the analysis of new sequences of species from fish hosts. Parasitology Research, 113(12), 4327-4333. [CrossRef] [PubMed] [Google Scholar]
- Li M, Wang J, Zhang J, Gu Z, Ling F, Ke X, Gong X. 2008. First report of two Balantidium species from the Chinese giant salamander, Andrias davidianus: Balantidium sinensis Nie 1935 and Balantidium andianusis n. sp. Parasitology Research, 102(4), 605-611. [CrossRef] [PubMed] [Google Scholar]
- Lynn D. 2008. The ciliated protozoa: characterization, classification, and guide to the literature. Dordrecht: Springer Science & Business Media. p. 373. [Google Scholar]
- Mahoon MS, Khan MI. 1986. Entozoic protozoa of frog Rana cyanophlyctis Schneider. Biologia (Lahore), 32(32), 383-420. [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene, 71(2), 491-499. [CrossRef] [Google Scholar]
- Nie D. 1932. Intestinal ciliates of Amphibia of Nanking. Science Society of China: Nanking. [Google Scholar]
- Pal NL, Dasgupta B. 1978. Observations on two new species of Balantidium in the Indian salamander, Tylototriton verrucosus (Caudata: Salamandridae). Proceedings of the Zoological Society, 31, 47-52. [Google Scholar]
- Pomajbíková K, Oborník M, Horák A, Petrželková KJ, Grim JN, Levecke B, Todd A, Mulama M, Kiyang J, Modrý D. 2013. Novel insights into the genetic diversity of Balantidium and Balantidium-like cyst-forming ciliates. PLoS Neglected Tropical Diseases, 7(3), e2140. [CrossRef] [PubMed] [Google Scholar]
- Ponce-Gordo F, Jimenez-Ruiz E, Martinez-Diaz R. 2008. Tentative identification of the species of Balantidium from ostriches (Struthio camelus) as Balantidium coli-like by analysis of polymorphic DNA. Veterinary Parasitology, 157(1), 41-49. [CrossRef] [PubMed] [Google Scholar]
- Puytorac PD, Grain J. 1965. Structure et ultrastructure de Balantidium xenopi sp. nov. Cilié trichostome parasite du batracien Xenopus fraseri Boul. Protistologica, 1, 29-36. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research, 41(D1), D590-D596. [CrossRef] [Google Scholar]
- Ray H. 1932. On the morphology of Balantidium sushilii n. sp., from Rana Tigrina Daud. Journal of Microscopy, 52(4), 374-382. [Google Scholar]
- Rodriguez JM. 1939. On the morphology of Balantidium kirbyi n. sp., from the Plathander. Journal of Parasitology, 25(3), 197-201. [CrossRef] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3), 539-542. [CrossRef] [PubMed] [Google Scholar]
- Şenler NGL, Yildiz İ. 2000. The ciliate fauna in the digestive system of Rana ridibunda (Amphibia: Anura) I: Balantidium (Balantidiidae, Trichostomatida). Turkish Journal of Zoology, 24(1), 33-44. [Google Scholar]
- Shete SG, Krishnamurthy R. 1984. Observations on the rectal ciliates of the genus Balantidium, Claparede and Lachmann, 1858 from Indian amphibians Rana tigrina and R. cyanophlyctis. Archiv für Protistenkunde, 128(1-2), 179-194. [CrossRef] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9), 1312-1313. [CrossRef] [PubMed] [Google Scholar]
- Stein F. 1867. Der Organismus der Infusionsthiere nach eigenen Forschungen in systematischer Reihenfolge bearbeitet: Leipzig. [Google Scholar]
- Strüder-Kypke MC, Kornilova OA, Lynn DH. 2007. Phylogeny of trichostome ciliates (Ciliophora, Litostomatea) endosymbiotic in the Yakut horse (Equus caballus). European Journal of Protistology, 43(4), 319-328. [CrossRef] [PubMed] [Google Scholar]
- Strüder-Kypke MC, Wright A-DG, Foissner W, Chatzinotas A, Lynn DH. 2006. Molecular phylogeny of litostome ciliates (Ciliophora, Litostomatea) with emphasis on free-living haptorian genera. Protist, 157(3), 261-278. [CrossRef] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725-2729. [CrossRef] [PubMed] [Google Scholar]
- Walker EL. 1909. Sporulation in the parasitic Ciliata. Archiv für Protistenkunde, 17, 297. [Google Scholar]
- Wright A-DG, Lynn DH. 1997. Monophyly of the trichostome ciliates (Phylum Ciliophora: Class Litostomatea) tested using new 18S rRNA sequences from the vestibuliferids, Isotricha intestinalis and Dasytricha ruminantium, and the haptorian, Didinium nasutum. European Journal of Protistology, 33(3), 305-315. [CrossRef] [Google Scholar]
- Wright A-DG, Lynn DH. 1997. Phylogenetic analysis of the rumen ciliate family Ophryoscolecidae based on 18S ribosomal RNA sequences, with new sequences from Diplodinium, Eudiplodinium, and Ophryoscolex. Canadian Journal of Zoology, 75(6), 963-970. [CrossRef] [Google Scholar]
- Wright ADG, Dehority BA, Lynn DH. 1997. Phylogeny of the rumen ciliates Entodinium, Epidinium and Polyplastron (Litostomatea: Entodiniomorphida) inferred from small subunit ribosomal RNA sequences. Journal of Eukaryotic Microbiology, 44(1), 61-67. [CrossRef] [Google Scholar]
Cite this article as: Zhao W, Li C, Zhang D, Wang R, Zheng Y, Zou H, Li W, Wu S, Wang G, Li M. 2018. Balantidium grimi n. sp. (Ciliophora, Litostomatea), a new species inhabiting the rectum of the frog Quasipaa spinosa from Lishui, China. Parasite 25, 29
All Tables
Comparison of body length (Lb), vestibulum length (Lv) and the ratio of vestibulum length and body length (Lv/Lb) between B. grimi and four Balantidium species.
All Figures
 |
Figure 1 LM images of B. grimi. A. Specimens fixed in formalin (5%) and soaked in glycerine-alcohol (10%), showing the oval body shape, vestibulum (vb) and macronucleus (ma), a round contractile vacuole (cv) in the posterior and a cytoproct (cp) at the end of the body. Scale bar = 10 µm. B. Specimens fixed in formalin (5%) and soaked in glycerine-alcohol (10%), showing the long vestibulum (vb) surrounded by cilia. Scale bar = 10 µm. C-F. are protargol stained: C. showing the body shape, macronucleus (ma) and micronucleus (mi). Scale bar = 10 µm. D. showing the vestibulum and somatic kineties. Scale bar = 10 µm. E. showing the vestibulum (vb) and the oval macronucleus (ma) with a spherical micronucleus (mi) embedded in the middle. Scale bar = 10 µm. F. showing the relative position of macronucleus (ma) and micronucleus (mi). Scale bar = 5 µm. |
| In the text | |
 |
Figure 2 Schematic drawing of B. grimi, showing the general form and structures from the ventral-left view: vestibulum (vb), food particles (fp), macronucleus (ma), micronucleus (mi), contractile vacuole (cv) and cytoproct (cp). Scale bar = 10 µm. |
| In the text | |
 |
Figure 3 SEM images of B. grimi. A. Overview of the ventral-left side (oral side), showing the general form, vestibulum (arrow) and uniformly arranged cilia. Scale bar = 10 µm. B. Overview of the right side, showing the body surface is partially flattened and thickly ciliated. Scale bar = 10 µm. C. Ventral-left view of the “V”-shaped vestibulum (arrow). Scale bar = 5 µm. D. The left anterior area of ciliate, showing the vestibulum (vb), an interkinetal ridge (rd), the groove (gr) and the cilia (cl) extending from grooves and are close to one another. Scale bar = 5 µm. E. Selected enlargement of Figure 3D, showing a ridge (rd) between cilia. Scale bar = 2 µm. |
| In the text | |
 |
Figure 4 Phylogenetic relationships of the SSU-rRNA sequences of B. grimi marked in bold and other Trichostomatia species showing the position of B. grimi inferred by maximum likelihood method and Bayesian algorithm. The trees were rooted using the sequence of Nyctotheroides deslierresae as the outgroup taxa. Numbers at nodes indicate bootstrap percentage and posterior probability, respectively. The sequences corresponding to species of the genus Balantidium are shadowed. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


