| Issue |
Parasite
Volume 31, 2024
|
|
|---|---|---|
| Article Number | 15 | |
| Number of page(s) | 7 | |
| DOI | https://doi.org/10.1051/parasite/2024009 | |
| Published online | 22 March 2024 | |
Research Article
Trypanosoma brucei gambiense group 2 experimental in vivo life cycle: from procyclic to bloodstream form
Cycle de vie expérimental in vivo de Trypanosoma brucei gambiense groupe 2 : de la forme procyclique à la forme sanguicole
INTERTRYP, Université de Montpellier, Cirad, IRD, Montpellier, France
* Corresponding author: sophie.ravel@ird.fr
Received:
27
October
2023
Accepted:
6
February
2024
Trypanosoma brucei gambiense (Tbg) group 2 is a subgroup of trypanosomes able to infect humans and is found in West and Central Africa. Unlike other agents causing sleeping sickness, such as Tbg group 1 and Trypanosoma brucei rhodesiense, Tbg2 lacks the typical molecular markers associated with resistance to human serum. Only 36 strains of Tbg2 have been documented, and therefore, very limited research has been conducted despite their zoonotic nature. Some of these strains are only available in their procyclic form, which hinders human serum resistance assays and mechanistic studies. Furthermore, the understanding of Tbg2’s potential to infect tsetse flies and mammalian hosts is limited. In this study, 165 Glossina palpalis gambiensis flies were experimentally infected with procyclic Tbg2 parasites. It was found that 35 days post-infection, 43 flies out of the 80 still alive were found to be Tbg2 PCR-positive in the saliva. These flies were able to infect 3 out of the 4 mice used for blood-feeding. Dissection revealed that only six flies in fact carried mature infections in their midguts and salivary glands. Importantly, a single fly with a mature infection was sufficient to infect a mammalian host. This Tbg2 transmission success confirms that Tbg2 strains can establish in tsetse flies and infect mammalian hosts. This study describes an effective in vivo protocol for transforming Tbg2 from procyclic to bloodstream form, reproducing the complete Tbg2 cycle from G. p. gambiensis to mice. These findings provide valuable insights into Tbg2’s host infectivity, and will facilitate further research on mechanisms of human serum resistance.
Résumé
Trypanosoma brucei gambiense (Tbg) groupe 2 est un sous-groupe de trypanosomes capables d’infecter l’Homme, présent en Afrique de l’Ouest et en Afrique centrale. Contrairement aux autres agents responsables de la maladie du sommeil, tels que Tbg groupe 1 et Trypanosoma brucei rhodesiense, Tbg2 ne présente pas les marqueurs moléculaires habituellement associés à la résistance au sérum humain. Seules trente-six souches de Tbg2 ont été répertoriées, limitant considérablement les recherches sur ce sous-groupe malgré sa nature zoonotique. Certaines de ces souches ne sont disponibles que sous leur forme procyclique, ce qui freine la réalisation des tests de résistance au sérum humain et les études mécanistiques. De plus, la compréhension du potentiel de Tbg2 à infecter les glossines et les hôtes mammifères est limitée. Dans cette étude, 165 glossines Glossina palpalis gambiensis ont été infectées expérimentalement par des parasites Tbg2 sous leur forme procyclique. Trente-cinq jours après l’infection, 43 des 80 glossines encore en vie se sont révélées positives à Tbg2 en PCR sur leur salive. Ces glossines ont réussi à infecter trois des quatre souris utilisées pour leur repas de sang. La dissection des glossines a révélé que seules six d’entre elles étaient réellement porteuses d’infections matures dans leur intestin et leurs glandes salivaires. Il est important de noter qu’une seule glossine porteuse d’une infection mature a suffi pour infecter un hôte mammifère. Ce succès de transmission de Tbg2 confirme que les souches de Tbg2 peuvent s’établir dans les glossines et infecter des hôtes mammifères. Cette étude décrit un protocole in vivo pour transformer la forme procyclique de Tbg2 en forme sanguicole, en reproduisant le cycle complet de Tbg2 de G. p. gambiensis à la souris. Ces résultats fournissent des informations précieuses sur le potentiel infectieux de Tbg2 et faciliteront la recherche sur les mécanismes de résistance au sérum humain des souches.
Key words: Trypanosoma brucei gambiense / Procyclic form / Bloodstream form / Glossina / Life cycle
© P. Juban et al., published by EDP Sciences, 2024
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Trypanosoma brucei (Tb) is an extracellular protozoan parasite transmitted by an arthropod hematophagous vector: the tsetse fly (Glossina spp.) [16]. Among the Tb species, the Tb brucei (Tbb) sub-species causes animal African trypanosomiasis or nagana in fauna. The human form of the disease, human African trypanosomiasis (HAT) or sleeping sickness is caused by the other two Tb sub-species: Tb rhodesiense (Tbr) and Tb gambiense (Tbg) [6]. Tbr causes an acute form of the disease in East Africa, whereas Tbg develops into a chronic form in Central and West Africa. Tbg HAT was responsible of 87% of the reported cases in 2019–2020 and is targeted by the World Health Organization for interruption of transmission by 2030 [9]. During the 1980s, the development of new analytical molecular methods allowed for the division of the Tbg subspecies into two groups. The most prevalent, genetically homogenous and monophyletic was group 1 (Tbg1) [11], invariably resistant to normal human serum (NHS) particularly thanks to the expression of the Tbg-specific glycoprotein (TgsGP) [2, 29]. In a recent review, group 2 (Tbg2) was defined as all human-infective Tb trypanosomes from West and Central Africa that do not fit into Tbg1 using various molecular markers [17]. Tbg2, Tbb, and Tbr are obviously highly diverse lineages but Tbg2 is different from Tbr with a consistent lack of serum resistance associated gene (SRA) [12]. If Tbg2 and its inconsistent resistance to lysis by human serum [21] does not appear to be a public health problem with only 36 strains referenced in the literature regarding the above definition, it represents a zoonotic form of HAT with a risk of transmission from animals to humans. In the current elimination context, it seems crucial to be able to detect such infections using adapted effective diagnosis and to determine if they are due to human serum resistance (HSR) trypanosomes or patient immunodeficiency (constitutive or transient) in order to implement adapted control strategies. Tbg2 stocks from different laboratories were gathered at UMR INTERTRYP (IRD/CIRAD, Montpellier, France) to study the HSR mechanisms and provide essential elements to anticipate the appearance of new mechanisms and to prevent possible phenomena of emergence [17].
Tb parasites have a multistage life cycle divided between the tsetse fly vector and a mammalian host. Along this life cycle, the parasite should continuously adapt to its surrounding environment. In the mammalian host, the bloodstream form (BSF) trypanosomes exist either in a proliferative long slender form or in a quiescent short stumpy form pre-adapted to the vector (Supplementary file 1). Following the infective blood meal, trypanosomes transform into their replicative procyclic form (PCF) in the tsetse fly midgut. Approximately one month after the infective meal, in a small proportion of tsetse flies (about 0.01% in natural conditions), trypanosomes colonize the salivary glands where they attach as epimastigote forms (EMF) [10]. Trypanosomes finally differentiate into infectious metacyclic forms (MCF) that can be transmitted to the mammalian host during the next blood meal.
Most of the Tbg2 strains are only available in their PCF and cannot be tested for their resistance to NHS, and for the study of the mechanisms implied. Moreover, very little is known about the potential for infection of Tbg2 in tsetse and mammalian hosts. Some rare studies have been conducted using PCF of Tbg1 or Tbg2, but transmissions to a mammalian host were not successful or were not attempted [25, 27]. The objectives of the present experimental study were (i) to confirm that Tbg2 PCF can settle in tsetse flies and become infectious to mice, and (ii) to transform Tbg2 PCF to BSF for further studies on HSR.
Material and methods
Ethics for animal experiments
Mice were kept under strict ethical conditions according to the international guidelines for the care and use of laboratory animals. The experiments designed for this study were approved by the Regional Ethics Committee for Animal Experimentation CEEA-LR 36 under project number APAFiS #34149 and authorized by the French Ministry for Higher Education and Research.
Tsetse flies
In this study, tsetse flies were used from a colony of G. p. gambiensis originating from Burkina Faso and maintained at CIRAD (Montpellier, France). Only males were chosen, as they develop a higher proportion of salivary gland infections (mature infections) compared to females when experimentally infected with trypanosomes of the subgenus Trypanozoon [8, 19, 22]. Due to several issues relative to fly physiology (natural death, fluctuating infecting meal feedings, low rate of trypanosome colonization in the midgut and of mature infection in the salivary glands) already described elsewhere [20, 23], 165 tsetse flies were used assuming that this number would be sufficient to obtain mature infections after one month. Before the infection process, one wing was removed from each fly for security reason.
Trypanosomes
The Tbg2 HTAG107-1 strain, also known as IPR107-1, was used in this study. This strain was isolated from humans in 1986 in the Daloa HAT focus, Côte d’Ivoire [28]. HTAG107-1 was grown in SDM-79 medium [3] supplemented with 20% fetal calf serum previously decomplemented (30 minutes at 56 °C). PCF trypanosomes were cultivated at 28 °C and harvested when 2.4 × 108 parasites were obtained (1 × 107 p/mL) (Figure 1). Parasites were then collected after centrifugation (1500×g, 10 minutes) and the pellet containing trypanosomes was washed three times with phosphate-buffered saline (PBS 1X) before use.
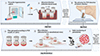 |
Figure 1 Trypanosoma brucei gambiense group 2 experimental in vivo cycle protocol (pi: post infection). +++: strong PCR signal; ++: medium PCR signal; +: weak PCR signal. PSG: phosphate-buffered saline-glucose. |
Experimental infection of tsetse flies
PCF trypanosomes (2.4 × 108) were gently mixed with 40 mL of sheep blood heated to 31 °C. The infected blood was proposed to starved G. p. gambiensis teneral males through a silicone membrane [1]. After the infective meal, tsetse flies were separated according to their blood-feeder status (blood meal visible in the abdomen or not). After 24 h, the process of infective meal was repeated with the non-gorged tsetse flies to ensure maximum infection rate success. Flies were then fed with uninfected sheep blood, three days a week for 35 days.
Salivation assay and PCR
Thirty-five days after the infective meal (sufficient time to obtain mature infection in the salivary glands [25, 30]), surviving tsetse flies were individually subjected to a salivation test. Each fly was allowed to salivate into a drop of phosphate-buffered saline–glucose (PSG) on warmed slides (37 °C) [4, 14] and immediately placed into an individual cage. The salivate was recovered from the slide in 25 μL of sterile water (Figure 1). To identify flies whose saliva carried trypanosomes, DNA was extracted from each collected saliva and analyzed by TBR1/2 PCR, as already described [25]. Flies with PCR-positive saliva were then grouped in different cages according to PCR signal intensity (strong, medium, or weak) (Figure 1). Flies with PCR-negative saliva and flies whose salivate could not be recovered because flies refused to salivate were euthanized and dissected for microscopic observation (×400).
Monitoring and dissection of the tsetse flies
Midguts of all flies found dead during the process were dissected from day 5 (time needed to observe parasite colonization of the midgut) to day 19 post-infection (pi). From day 20 pi, the salivary glands were also dissected (assuming that no trypanosomes can be found in the salivary glands before this time). All the dissected midguts and salivary glands were examined for trypanosomes by phase contrast microscopy at 400× magnification.
Infection of mice
At day 39 and 42 pi, each group of flies with PCR-positive saliva was fed twice (3 days apart) on the belly of anesthetized female BALB/c mice previously immuno-suppressed with 0.15 mL of cyclophosphamide (ENDOXAN, 20 mg/mL) injected subcutaneously (Figure 1). A different mouse was assigned to each group of flies. The objective of this sorting was to maximize the success of infection of one of the mice. Tsetse were starved for three days prior to the mice blood meal to increase sting probability. The parasitemia of the mice was then determined daily by microscopy using the rapid “matching” method [15] on a drop of blood collected from the tail of each mouse (Figure 1).
The mice-fed surviving flies were euthanized and dissected for microscopic (×400) observation of the midgut and salivary glands.
Results and discussion
Molecular screening of tsetse flies with trypanosomes in their salivary glands
Thirty-five days after tsetse fly infection, 80 flies out of 165 were still alive (Figure 2). Out of them, 78 were tested for the detection of trypanosomes in saliva by PCR and 43 (55%) showed PCR-positive saliva. Out of these 43 flies, 9, 12, and 22 exhibited a strong, medium, and weak PCR signal, respectively (Supplementary file 2). Between the beginning of the collection of the saliva and the results of the PCR analysis, 8 flies died. The remaining 35 flies were grouped into four cages: two containing 9 flies each with weak PCR signal, one containing 11 flies with medium PCR signal, and one containing 6 flies with strong PCR signal (Figure 2). All the PCR-negative flies were euthanized and dissected for microscopic observation of the midgut and salivary glands.
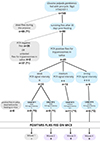 |
Figure 2 Diagram summarizing the main results of the experimental infection of flies and mice with Trypanosoma brucei gambiense group 2 HTAG-107 strain. (*x: number of flies found positive in salivary glands by microscopic observation after dissection). |
Monitoring of tsetse fly infection
Throughout this experiment, 115 flies could be dissected of which 11 (9.6%) showed parasites in their midgut only, and six (5.2%) in both their midgut and salivary glands. Among the flies showing trypanosomes in their salivary glands that were fed on mice, none were found in cage No. 1, one was from cage No. 2, one from cage No. 3 and two from cage No. 4, in line with the molecular analysis.
The number of mature infections identified by the PCR analysis of the flies’ saliva (n = 43) was much higher than that determined by microscopic observation of the salivary glands (n = 6). These data are in the range found with previous observations from other studies (with maximum 10% flies found positive in SG) [25–27]. Because of its high sensitivity, PCR from saliva may offer a better view of mature infections as it is sometimes challenging to observe trypanosomes in salivary glands. However, for most of the PCR-positive flies for trypanosomes in their saliva, no trypanosomes were observed in the dissected salivary glands, despite diligent microscopic observation. Part of the PCR-positive saliva could result from flies only infected in the midgut, whose saliva may also contain regurgitated gut contents including trypanosome DNA.
Additionally, because the recovery of salivary glands is not easy during fly dissection, salivary glands may not always be available for observation. This was the case for three of the flies dissected at day 36 pi: trypanosomes were observed in the midgut only, but the salivary glands could not be recovered for technical reasons. Therefore, the percentage of flies with mature infection may have been underestimated through microscopy techniques.
Monitoring infection in mice
Once the trypanosome PCR-positive flies from the four cages had been fed on mice, parasitemia was monitored daily. Three mice out of the four developed an infection two to four days after the blood meal (Figure 2 and Supplementary file 3). The resulting HTAG107-1 BSF trypanosomes were collected from the infected mice by cardiac puncture and supplemented with 50% glycerol before being stored in liquid nitrogen. Dissection and observation of the salivary glands of the flies used to bite the mice showed that two flies were positive for trypanosomes in cage No. 4, one in cage No. 3, and one in cage No. 2. No salivary gland-positive flies were detected in cage No. 1, which is congruent with the absence of infection in mouse 1.
Advantages and drawbacks to reconstitute a PCF to BSF experimental in vivo life cycle
From 165 flies fed with a single meal of sheep blood mixed with cultivated PCF trypanosomes, at least 6 flies with mature infections after one month were obtained.
We succeeded in infecting mice from infected tsetse flies. This achievement is partly due to the large size of the starting sample (n = 165) and to the starvation of the flies before the infective meal. Post-dissection of the flies used to infect the mice showed that a mouse only needs to be bitten by one fly with mature infection to become infected. This success also confirms that Tbg2 group strains can settle in the tsetse fly and infect a mammalian host.
For strains only available as PCF, the results of this experiment made it possible to obtain the bloodstream form that can be evaluated for resistance to NHS. However, this experiment is time consuming, requires great technical effort and is not in line with current animal ethics principles (3R rule – replace, reduce, refine). For this reason, if the passage from PCF to BSF is the only result desired, in vitro plasmid methods should be preferred [24].
While HAT elimination seems a realistic goal for 2030, we advocate for improving knowledge of the Tbg strains that are still circulating, even at a low, almost undetectable level. In several HAT foci, parasitemia observed in human or in animal reservoirs is very low [7] and hinders deep genotypical and phenotypical characterization. Isolating strains and mastering a transmission cycle makes it possible to collect data that are not available otherwise, for instance to account for differences in pathogenicity and virulence to humans over natural cycles [5]. This is particularly interesting in the case of Tbg2 strains. They have been found to be resistant or partially resistant to the NHS, but their ability to maintain NHS resistance capacity after cycling in animals is unknown. Deciphering the nature of the resistance to NHS – constitutive (as is the case for Tbg1) or conditional (which can be lost after several vector/animal cycles) – is keystone information for the effective and sustainable elimination of sleeping sickness. Indeed, elimination of HAT will lead to a decrease in acquired immunity in populations, which could create major concerns for more susceptible populations if they are again exposed to strains with constitutive resistance [17].
Finally, it remains difficult to reproduce complete cyclical transmission (from infection of the tsetse fly to transmission to the host by the tsetse fly) of Tbg2 and even Tbg1 because of the low rate of mature infections of Tbg [20]. Two other studies succeeded using clones of Tbg2 BSF and Glossina morsitans [13, 18]. In this study, we provide evidence of an effective in vivo protocol to transform Tbg2 PCF to BSF by experimentally reproducing the complete Tbg2 cycle from G. p. gambiensis to the mouse.
Acknowledgments
This work was supported by the IRD (Institut de Recherche pour le Développement). Paola Juban was supported by the KIM RIVE project (Key Initiative Muse Risks & Vectors) from Montpellier Université d’excellence (MUSE). All experiments on G. p. gambiensis were performed on the Baillarguet insectarium platform (https://doi.org/10.18167/infrastructure/00001), member of the National Infrastructure EMERG’IN and of the Vectopole Sud network (http://www.vectopole-sud.fr/). The Baillarguet insectarium platform is led by the joint units Intertryp (IRD, Cirad) and ASTRE (Cirad, INRAE). Figure 1 and Supplementary files 1 and 3 are original figures created with Biorender.com. We would like to thank Bernadette Tchicaya and Annick Ranaivoarisoa for providing males of G. p. gambiensis.
Conflict of interests
The authors declare that they have no competing interests.
Supplementary material
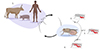 |
Supplementary file 1: Schematic life cycle of Trypanosoma brucei. 1: Bloodstream trypanosomes in mammalian host; A: long slender form; B: short stumpy form. 2: Procyclic trypanosomes in tsetse fly midgut. 3: Epimastigote trypanosomes colonize salivary glands. 4: Metacyclic trypanosomes in salivary glands can be transmitted to a mammalian host during the next tsetse fly meal. In red: localization of the parasites in the tsetse fly through the cycle. |
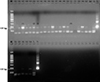 |
Supplementary file 2: TBR1/2 PCR identification of Trypanosoma brucei s.l in tsetse fly saliva showing 177 bp DNA satellite repeat specific for T. brucei s.l – numbers correspond to those assigned to the flies. M: 100 bp DNA size marker; Tex: DNA extraction negative control; T−: PCR-negative control; T+: positive control; (1): Strong PCR signal; (2): Medium PCR signal; (3): Weak PCR signal; (4): Negative PCR signal. |
Supplementary file 3: Microscopic observation videos of Trypanosoma brucei gambiense group 2 throughout its life cycle, including (1) procyclic form in tsetse midgut, (2) metacyclic form in tsetse salivary glands, and (3) bloodstream form found in infected mice blood. Access here
References
- Bauer B, Wetzel H. 1976. A new membrane for feeding Glossina morsitans Westw. (Diptera, Glossinidae). Bulletin of Entomological Research, 65(4), 563–565. [CrossRef] [Google Scholar]
- Berberof M, Pérez-Morga D, Pays E. 2001. A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Molecular and Biochemical Parasitology, 113(1), 127–138. [CrossRef] [PubMed] [Google Scholar]
- Brun R, Schönenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta tropica, 36(3), 289–292. [PubMed] [Google Scholar]
- Burtt E. 1946. Salivation by Glossina morsitans on to glass slides: a technique for isolating infected flies. Annals of Tropical Medicine and Parasitology, 40(2), 141–144. [CrossRef] [PubMed] [Google Scholar]
- Büscher P, Bart JM, Boelaert M, Bucheton B, Cecchi G, Chitnis N, Courtin D, Figueiredo LM, Franco JR, Grébaut P, Hasker E, Ilboudo H, Jamonneau V, Koffi M, Lejon V, MacLeod A, Masumu J, Matovu E, Mattioli R, Noyes H, Picado A, Rock KS, Rotureau B, Simo G, Thévenon S, Trindade S, Truc P, Van Reet N. 2018. Do cryptic reservoirs threaten gambiense-sleeping sickness elimination? Trends in Parasitology, 34(3), 197–207. [CrossRef] [PubMed] [Google Scholar]
- Büscher P, Cecchi G, Jamonneau V, Priotto G. 2017. Human African trypanosomiasis. Lancet, 390, 2397–2409. [CrossRef] [PubMed] [Google Scholar]
- Camara M, Camara O, Ilboudo H, Sakande H, Kaboré J, N’Dri L, Jamonneau V, Bucheton B. 2010. Sleeping sickness diagnosis: use of buffy coats improves the sensitivity of the mini anion exchange centrifugation test. Tropical Medicine & International Health, 15(7), 796–799. [CrossRef] [PubMed] [Google Scholar]
- Dale C, Welburn SC, Maudlin I, Milligan PJ. 1995. The kinetics of maturation of trypanosome infections in tsetse. Parasitology, 111(2), 187–191. [CrossRef] [PubMed] [Google Scholar]
- Franco JR, Cecchi G, Paone M, Diarra A, Grout L, Kadima Ebeja A, Simarro PP, Zhao W, Argaw D. 2022. The elimination of human African trypanosomiasis: Achievements in relation to WHO road map targets for 2020. PLoS Neglected Tropical Diseases, 16(1), e0010047. [CrossRef] [PubMed] [Google Scholar]
- Frézil J, Cuisance D. 1994. Trypanosomiases, maladies d’avenir : leurs perspectives et leurs inconnues. Trypanosomiasis, diseases with future: prospects and uncertainty. Bulletin de la Société de Pathologie Exotique, 87, 391–393. [Google Scholar]
- Gibson WC. 1986. Will the real Trypanosoma brucei gambiense please stand up. Parasitology Today, 2(9), 255–257. [CrossRef] [Google Scholar]
- Gibson W, Backhouse T, Griffiths A. 2002. The human serum resistance associated gene is ubiquitous and conserved in Trypanosoma brucei rhodesiense throughout East Africa. Infection, Genetics and Evolution, 1(3), 207–214. [CrossRef] [PubMed] [Google Scholar]
- Gibson W, Winters K, Mizen G, Kearns J, Bailey M. 1997. Intraclonal mating in Trypanosoma brucei is associated with out-crossing. Microbiology, 143, 909–920. [CrossRef] [PubMed] [Google Scholar]
- Gidudu AM, Cuisance D, Reifenberg JM, Frézil JL. 1995. Amélioration de la technique de salivation des glossines pour la détection des métatrypanosomes infectants : étude de quelques facteurs biologiques et non biologiques sur le comportement de sondage des glossines. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 48(2), 153–160. [Google Scholar]
- Herbert WJ, Lumsden WHR. 1976. Trypanosoma brucei: a rapid “matching” method for estimating the host’s parasitemia. Experimental Parasitology, 40(3), 427–431. [CrossRef] [PubMed] [Google Scholar]
- Hoare CA. 1972. The trypanosomes of mammals. A zoological monograph. Blackwell Scientific Publications: Oxford, UK. [Google Scholar]
- Jamonneau V, Truc P, Grébaut P, Herder S, Ravel S, Solano P, De Meeus T. 2019. Trypanosoma brucei gambiense Group 2: the unusual suspect. Trends in Parasitology, 35(12), 983–995. [CrossRef] [PubMed] [Google Scholar]
- Jenni L, Marti S, Schweizer J, Betschart B, Le Page RWF, Wells JM, Tait A, Paindavoine P, Pays E, Steinert M. 1986. Hybrid formation between African trypanosomes during cyclical transmission. Nature, 322, 173–175. [CrossRef] [PubMed] [Google Scholar]
- Maudlin I, Welburn SC, Milligan PJ. 1991. Salivary gland infection: a sex-linked recessive character in tsetse? Acta Tropica, 48(1), 9–15. [Google Scholar]
- Maudlin I, Welburn SC. 1994. Maturation of trypanosomes infection in tsetse. Experimental Parasitology, 79, 202–205. [CrossRef] [PubMed] [Google Scholar]
- Mehlitz D. 1986. Le réservoir animal de la maladie du sommeil à Trypanosoma brucei gambiense. CIRAD-IEMVT: Maisons-Alfort, 167 p. (Etudes et synthèses de l’IEMVT, n. 18) ISBN 2-85985-127-5. [Google Scholar]
- Milligan PJ, Maudlin I, Welburn SC. 1995. Trypanozoon: infectivity to humans is linked to reduced transmissibility in tsetse: II. Genetic mechanisms. Experimental Parasitology, 81(3), 409–415. [CrossRef] [PubMed] [Google Scholar]
- Moloo SK, Asonganyi T, Jenni L. 1986. Cyclical development of Trypanosoma brucei gambiense from cattle and goats in Glossina. Acta Tropica, 43, 407–408. [PubMed] [Google Scholar]
- Mugo E, Egler F, Clayton C. 2017. Conversion of procyclic-form Trypanosoma brucei to the bloodstream form by transient expression of RBP10. Molecular and Biochemical Parasitology, 216, 49–51. [CrossRef] [PubMed] [Google Scholar]
- Ravel S, Grébaut P, Cuisance D, Cuny G. 2003. Monitoring the developmental status of Trypanosoma brucei gambiense in the tsetse fly by means of PCR analysis of anal and saliva drops. Acta Tropica, 88(2), 161–165. [CrossRef] [PubMed] [Google Scholar]
- Ravel S, Patrel D, Koffi M, Jamonneau V, Cuny G. 2006. Cyclical transmission of Trypanosoma brucei gambiense in Glossina palpalis gambiensis displays great differences among field isolates. Acta Tropica, 100(1–2), 151–155. [CrossRef] [PubMed] [Google Scholar]
- Richner D, Brun R, Jenni L. 1988. Production of metacyclic forms by cyclical transmission of West African Trypanosoma (T.) brucei isolates from man and animals. Acta Tropica, 45(4), 309–319. [PubMed] [Google Scholar]
- Stevens JR, Lanham SM, Allingham R, Gashumba JK. 1992. A simplified method for identifying subspecies and strain groups in Trypanozoon by isoenzymes. Annals of Tropical Medicine and Parasitology, 86(1), 9–28. [CrossRef] [PubMed] [Google Scholar]
- Uzureau P, Uzureau S, Lecordier L, Fontaine F, Tebabi P, Homblé F, Grélard A, Zhendre V, Nolan DP, Lins L, Crower JM, Pays A, Felu C, Poelvoorde P, Vanhollebeke B, Moestrup SK, Lyngso J, Pedersen JS, Mottram JC, Dufourc EJ, Morga DP, Pays E. 2013. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature, 501(7467), 430–434. [CrossRef] [PubMed] [Google Scholar]
- Van Den Abbeele J, Claes Y, Van Bickstaele D, Le Ray D, Coosemans M. 1999. Trypanosoma brucei spp. development in the tsetse fly: Characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology, 118, 469–478. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Juban P, Bart J-M, Ségard A, Jamonneau V & Ravel S. 2024. Trypanosoma brucei gambiense group 2 experimental in vivo life cycle: from procyclic to bloodstream form. Parasite 31, 15.
All Figures
 |
Figure 1 Trypanosoma brucei gambiense group 2 experimental in vivo cycle protocol (pi: post infection). +++: strong PCR signal; ++: medium PCR signal; +: weak PCR signal. PSG: phosphate-buffered saline-glucose. |
| In the text | |
 |
Figure 2 Diagram summarizing the main results of the experimental infection of flies and mice with Trypanosoma brucei gambiense group 2 HTAG-107 strain. (*x: number of flies found positive in salivary glands by microscopic observation after dissection). |
| In the text | |
 |
Supplementary file 1: Schematic life cycle of Trypanosoma brucei. 1: Bloodstream trypanosomes in mammalian host; A: long slender form; B: short stumpy form. 2: Procyclic trypanosomes in tsetse fly midgut. 3: Epimastigote trypanosomes colonize salivary glands. 4: Metacyclic trypanosomes in salivary glands can be transmitted to a mammalian host during the next tsetse fly meal. In red: localization of the parasites in the tsetse fly through the cycle. |
| In the text | |
 |
Supplementary file 2: TBR1/2 PCR identification of Trypanosoma brucei s.l in tsetse fly saliva showing 177 bp DNA satellite repeat specific for T. brucei s.l – numbers correspond to those assigned to the flies. M: 100 bp DNA size marker; Tex: DNA extraction negative control; T−: PCR-negative control; T+: positive control; (1): Strong PCR signal; (2): Medium PCR signal; (3): Weak PCR signal; (4): Negative PCR signal. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


