| Issue |
Parasite
Volume 30, 2023
|
|
|---|---|---|
| Article Number | 39 | |
| Number of page(s) | 8 | |
| DOI | https://doi.org/10.1051/parasite/2023044 | |
| Published online | 27 September 2023 | |
Research Article
High genotype diversity and zoonotic potential of Enterocytozoon bieneusi in yaks (Bos grunniens) from Ganzi Tibetan Autonomous Prefecture, Sichuan Province
Grande diversité génotypique et potentiel zoonotique d’Enterocytozoon bieneusi chez les yaks (Bos grunniens) de la préfecture autonome tibétaine de Ganzi, province du Sichuan
1
College of Veterinary Medicine, Northwest A&F University, Yangling 712100, China
2
Animal Husbandry Science Institute of Ganzi Tibetan Autonomous Prefecture, Kangding 626000, China
3
College of Veterinary Medicine, Huazhong Agricultural University, Wuhan 430070, China
4
Engineering Research Center of Efficient New Vaccines for Animals, Ministry of Education, Yangling 712100, China
5
Key Laboratory of Ruminant Disease Prevention and Control (West), Ministry of Agriculture and Rural Affairs, Yangling 712100, China
6
Engineering Research Center of Efficient New Vaccines for Animals, Universities of Shaanxi Province, Yangling 712100, China
* Corresponding author: zgh083@nwsuaf.edu.cn
Received:
17
June
2023
Accepted:
3
September
2023
Enterocytozoon bieneusi is a common pathogen in humans and various animals, threatening the breeding industry and public health. However, there is limited information on the molecular characteristics of E. bieneusi in yaks, an economically important animal mainly domesticated in the Qinghai Tibet Plateau in China. In the present study, nested PCR targeting the ITS gene region was applied to investigate the positive rates and genetic diversity of E. bieneusi in 223 faecal samples of yaks from three locations in Ganzi Tibetan Autonomous Prefecture, Sichuan Province. The total positive rate of E. bieneusi was 23.8% (53/223). Significant differences in positive rates were identified among yaks from three locations (χ2 = 8.535, p = 0.014) and four age groups (χ2 = 17.259, p = 0.001), with the highest positive rates in yaks from Yajiang and aged < 6 months, respectively. Sequence analysis identified seven known (EbpC, LW1, LQ10, PigEBITS5, ESH-01, J and BEB4) and five novel (Ganzi1–5) ITS genotypes. Phylogenetic analysis showed eight genotypes (EbpC, LW1, LQ10, PigEBITS5, ESH-01, Ganzi1, Ganzi2 and Ganzi4) in group 1 and three genotypes (J, BEB4 and Ganzi3) in group 2, indicating high genotype diversity and zoonotic potential of E. bieneusi in yaks from Ganzi. Considering the increasing zoonotic genotypes in yaks in the present study compared with previous findings, interventions should be developed to reduce the potential transmission of E. bieneusi between humans and animals.
Résumé
Enterocytozoon bieneusi est un agent pathogène courant chez l'homme et chez divers animaux, menaçant l'industrie de l'élevage et la santé publique. Cependant, il existe peu d'informations sur les caractéristiques moléculaires d’E. bieneusi chez les yaks, un animal important pour l’économie, principalement domestiqué sur le plateau du Qinghai au Tibet en Chine. Dans la présente étude, une PCR imbriquée ciblant la région du gène ITS a été appliquée pour étudier la positivité et la diversité génétique d’E. bieneusi dans 223 échantillons fécaux de yaks provenant de trois sites de la préfecture autonome tibétaine de Ganzi, province du Sichuan. Le taux total de positivité pour E. bieneusi était de 23,8 % (53/223). Des différences significatives dans les taux positifs ont été identifiées parmi les yaks de trois emplacements (χ2 = 8,535, P = 0,014) et de quatre groupes d'âge (χ2 = 17,259, P = 0,001), avec les taux positifs les plus élevés respectivement chez les yaks de Yajiang et ceux âgés de moins de 6 mois. L'analyse de séquence a identifié sept génotypes ITS connus (EbpC, LW1, LQ10, PigEBITS5, ESH-01, J et BEB4) et cinq nouveaux (Ganzi1–5). L’analyse phylogénétique a montré huit génotypes (EbpC, LW1, LQ10, PigEBITS5, ESH-01, Ganzi1, Ganzi2 et Ganzi4) dans le groupe 1 et trois génotypes (J, BEB4 et Ganzi3) dans le groupe 2, indiquant une diversité génotypique élevée et un potentiel zoonotique d’E. bieneusi chez les yaks de Ganzi. Compte tenu de l’augmentation des génotypes zoonotiques chez les yaks dans la présente étude par rapport aux résultats précédents, des interventions devraient être développées pour réduire la transmission potentielle d’E. bieneusi entre les humains et les animaux.
Key words: Enterocytozoon bieneusi / Yak / Genotype / Zoonotic potential
© X. Yang et al., published by EDP Sciences, 2023
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Microsporidia are important opportunistic pathogens that lead to significant economic losses in animal breeding worldwide [11, 41]. Among them, Enterocytozoon bieneusi is one of the most common zoonotic species and contributes to over 90% of human cases of microsporidiosis [30, 41]. Although E. bieneusi usually causes asymptomatic infection in both immunocompetent and immunodeficient individuals, it can also lead to gastrointestinal disorders, wasting and diarrhoea, especially for immunocompromised populations (e.g., HIV patients) and children [10, 24, 34, 62]. Meanwhile, infected hosts can release mature spores that contaminate water and food, threatening public health [38, 52]. Thus, the Environmental Protection Agency and the National Institutes of Health of the United States have listed E. bieneusi as a microbial contaminant candidate for waterborne transmission and a Class B biodefense pathogen, respctively [4].
Knowledge of the distribution and genetic characterisation of pathogens can shed new light on the prevention and control of diseases. Based on the molecular characterisation of the ITS gene locus of E. bieneusi, over 600 genotypes from 11 genetic groups (groups 1–11) with divergent host specificity have been recognised [20, 21, 61]. Within group 1, a few genotypes, such as D, EbpC, Type IV, Peru6, Peru8 and Peru11, have been widely reported in both humans and animals, reflecting significant zoonotic importance of genotypes in this group [20]. Genotypes in group 2 were previously reported to be specific in ruminants, but expanding of the host range for some genotypes (e.g., BEB4, BEB6, I and J) indicates potential zoonotic significance or cross-species transmission capabilities within this group [20, 48, 49, 60]. Host specificity of genotypes was commonly found in the groups 3–11, reflected by the unique existence of WL6 in rodents, PtEb VIII in cats, and CAF4 in humans and non-human primates [1, 16, 20, 25, 43]. Most genotypes within groups 3–11 showed limited zoonotic potential [20]. However, due to the limited genotypes reported in the groups 3–11, further studies on more samples from diverse hosts and geographical areas are needed to verify zoonotic potential of genotypes within these groups.
Yaks are a unique livestock resource distributed in the Qinghai Tibet Plateau and its adjacent high and subalpine areas. These animals can adapt to harsh environments such as very low temperatures, hypoxia, and extreme dryness, and are an important and unique livestock species in production [55]. Under grazing condition, yaks can be easily infected with various parasitic pathogens, and some zoonotic pathogens have been reported in yaks, such as Cryptosporidium spp. [5], Echinococcus granulosus [22] and Toxoplasma gondii [46], indicating zoonotic potential of those pathogens.
Previous studies have reported positive rates of E. bieneusi in yaks from Gansu, Qinghai, Tibet and Yunnan in China, with positive rates ranging from 1.1% to 7.2%, and 15 genotypes, including five zoonotic genotypes (BEB4, BEB6, I, J and D) and ten animal-adapted genotypes (CHN11, CHN12, CHN13, CHN14, CHC8, CAM2, COS-I, NESH5, WCY1 and YAK1) [26, 29, 51, 58, 59]. To further explore the distribution of E. bieneusi in yaks, the present study investigated the positive rates and genotype distribution of E. bieneusi in yaks from three main breeding areas in Ganzi Tibetan Autonomous Prefecture, and assessed the zoonotic potential of this pathogen in yaks.
Materials and methods
Ethics statement
This study was conducted under the approval and instructions of the ethics committee of Northwest A&F University (DY2022048).
Sampling
This study was conducted in Ganzi Tibetan Autonomous Prefecture, Sichuan Province, China. Ganzi is located in the west of Sichuan Province and the southeastern Tibetan plateau (97°22′–102°29′ E, 27°58′–34°20′ N), with 153,000 km2 and an average of over 4,000 m above sea level. As an important economic livestock species in Ganzi, yaks are usually raised in separate enclosures and seldomly have opportunities for contact with other animals. From October 2022 to April 2023, a total of 223 faecal samples were collected from yaks in Seda (n = 66), Litang (n = 130) and Yajiang (n = 27) in Ganzi (Fig. 1). For each analysed population, animals lived in enclosures, with about 10–20 animals sharing the same enclosure, and usually did not have close contact with humans except for farm owners. During the time of sampling, animals analysed in the present study did not show obvious symptoms, except for diarrhoea of 40 animals. All the samples were collected directly from the rectum of animals, placed in separated bags, marked with sample information (e.g., location, age and sex), transported to the parasitology laboratory of Northwest A&F University under cool conditions as soon as possible, and preserved in 2.5% potassium dichromate under 4 °C.
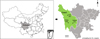 |
Figure 1 Geographical distribution of sampling sites in Ganzi Tibetan Autonomous Prefecture, Sichuan Province, China. |
Genomic DNA extraction
Faecal samples were washed in distilled water three times to remove potassium dichromate, and subsequently applied for total genomic DNA isolation using an E.Z.N.A. Stool DNA kit (Omega, Norcross, GA, USA), following the manufacturer’s instructions. The gDNA samples were stored at −20 °C.
PCR amplification
The colonisation frequency of E. bieneusi was detected using nested PCR based on the ITS gene locus, and using the primers previously reported [43]. A nested PCR was conducted in a 25 µL reaction mixture containing 1× Rapid Taq Master Mix, 0.4 µM each primer, 1 µL gDNA for the primary PCR or 1 µL primary PCR product for the secondary PCR, under the following conditions for both rounds: an initial denaturing at 94 °C for 5 min, followed by 35 cycles of 94 °C for 45 s, 55 °C for 45 s and 68 °C for 1 min, and a final extension at 68 °C for 7 min. A positive control with gDNA isolated from E. bieneusi-positive samples preserved in our laboratory and a negative control with distilled water were included in each PCR reaction. Positive secondary PCR conducts will show a band of ~392 bp under a UV transilluminator after 1% agarose gel electrophoresis.
Sequencing and sequence analysis
All positive secondary PCR products were sent to Sangon Biotech (Shanghai, China) for sequencing in both directions. The obtained sequences were identified to be E. bieneusi ITS gene by Blastn analysis at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). To assess the relationship of E. bieneusi genotypes found in the present study, a phylogenetic tree was developed using the maximum-likelihood (ML) method with the General Time Reversible model and bootstrap evaluation of 1000 replicates within MEGA V6.0 [45].
Statistical analysis
Differences in the positive rates of E. bieneusi in yaks among the location, age, sex and diarrhoea groups were analysed using a χ2 test in SPSS V18.0 (IBM, New York, NY, USA). Significant differences were confirmed if the p-value was less than 0.05.
Nucleotide sequence accession numbers
Representative nucleotide sequences of E. bieneusi ITS gene in the present study have been submitted to GenBank under accession numbers OR023607–OR023620.
Results
Occurrence of E. bieneusi in yaks
Of the 223 faecal samples examined in the present study, 53 (23.8%) were positive for E. bieneusi in yaks based on the PCR-sequencing tool targeting the ITS gene locus (Table 1). There were significant differences among the positive rates of E. bieneusi in three locations (χ2 = 8.535; p = 0.014), with the highest in Yajiang (44.4%, 12/27), followed by Seda (25.8%, 17/66) and Litang (18.5%, 24/130). Meanwhile, a significant difference in positive rates was also identified among four age groups (χ2 = 17.259; p = 0.001), with the highest in yaks aged <6 months (41.0%, 16/39), followed by 12–24 months (39.5%, 16/43), > 24 months (15.6%, 15/90) and 6–12 months (11.8%, 6/51). Although the positive rates of E. bieneusi varied among sex and diarrhoea groups, no significant differences were found (Table 1).
Occurrence, genotypes and factors associated with E. bieneusi infection in yaks from Ganzi Tibetan Autonomous Prefecture.
Distribution of E. bieneusi genotypes in yaks
Based on sequence analysis of the ITS gene locus of E. bieneusi, a total of 12 genotypes were identified in the 53 sequences in the present study, with seven known genotypes (BEB4, J, EbpC, LW1, LQ10, PigEBITS5 and ESH-01) and five novel genotypes (Ganzi1–5) (Table 1). No mixed infections of genotypes were identified in the present study. The novel genotypes identified in the present study have been re-sequenced, and the results indicated that these genotypes were truly novel. Ganzi1, Ganzi3 and Ganzi4 had three, one and two nucleotide substitutions compared with the genotypes EbpC (MN902235.1), BEB4 (MT231512.1) and CYG-1 (MZ479291.1), respectively. Ganzi2 had one nucleotide deletion compared with the genotype ESH-01 (KR902354.1). Ganzi5 had 31 nucleotide substitutions and two nucleotide deletions compared with the genotype XJH6 (MN704930.1). Among the identified genotypes, BEB4 was the commonest genotype found in 45.3% (24/53) of yak isolates, followed by J (22.6%, 12/53), EbpC (13.2%, 7/53), Ganzi4 (3.8%, 2/53) and other genotypes (1.9%, 1/53) (Table 1). There were sequence differences in genotype diversity among three locations, with six (BEB4, PigEBITS5, LQ10, Ganzi3–5), six (EbpC, BEB4, ESH-01, LW1, Ganzi1 and Ganzi2) and one (J) genotypes in Litang, Seda and Yajiang, respectively. Meanwhile, eight (BEB4, EbpC, ESH-01, LW1, PigEBITS5, LQ10, Ganzi2 and Ganzi3), three (J, BEB4 and Ganzi4), three (BEB4, J and Ganzi5) and three (BEB4, EbpC and Ganzi1) genotypes were identified in yaks aged >24 months, <6 months, 6–12 months and 12–24 months, respectively. More genotypes were found in female yaks (BEB4, PigEBITS5, Ganzi3 and Ganzi4) compared with male yaks (BEB4 and Ganzi5). A total of 11 genotypes were recognised in non-diarrhoea yaks (BEB4, J, EbpC, ESH-01, LW1, LQ10, Ganzi1–5), while only four were found in diarrhoeal yaks (BEB4, EbpC, PigEBITS5 and Ganzi4).
Phylogenetic relationships of E. bieneusi genotypes
Phylogenetic analysis based on the ITS gene locus of E. bieneusi indicated eight genotypes (EbpC, LW1, LQ10, PigEBITS5, ESH-01, Ganzi1, Ganzi2 and Ganzi4) contributing to 28.3% (15/53) of the isolates, and belonged to the potentially zoonotic group 1 (Fig. 2). Meanwhile, three genotypes (J, BEB4 and Ganzi3) belonged to group 2, with increasing zoonotic potential, while genotype Ganzi5 did not belong to any known group (Fig. 2).
 |
Figure 2 Phylogenetic relationships of E. bieneusi genotypes in this study, with reference sequences downloaded from GenBank based on the sequence analysis of the ITS locus using Maximum Likelihood analysis with the General Time Reversible model. Red and blue filled circles before the bold sample names represent novel and known genotypes identified in the present study, respectively. Bootstrap values (> 50) are indicated above the nodes. Scale bar indicates 0.1 nucleotide substitutions/site. Genotype SW3 from stormwater (KF591679.1) is used as the outgroup. |
Discussion
Enterocytozoon bieneusi is a common zoonotic pathogen threatening the health of humans and various animals [20]. Knowledge of the distribution and molecular genetics of pathogens could provide insights for the prevention and control of diseases. To further understand the colonisation frequency of E. bieneusi in bovine animals, the present study explored the occurrence and zoonotic potential of E. bieneusi in yaks from three locations in Ganzi Tibetan Autonomous Prefecture using PCR-sequencing targeted the ITS gene locus, and the results indicated high genetic diversity and zoonotic potential of E. bieneusi in yaks in this area.
Recently, E. bieneusi has been widely reported in bovine animals in China. In the present study, the positive rate of E. bieneusi in yaks was 23.8% (53/223), which was higher than that in yaks in Qinghai (7.0%; 7.2%) [26, 58], Gansu (1.13%) [29], and Tibet (5.0%) [51], water buffaloes in Jiangxi (5.6%) [19], Anhui (0.9%) [23] and Hunan (2.2%) [28], beef cattle in Jiangxi (3.9%) [19], Henan (5.4%) [28] and Shaanxi (19.7%) [50], and dairy cattle in most reported provinces in China, except for Heilongjiang (30.1%; 29.0%) [47, 63] and Jilin (37.6%) [60]. The disparities in positive rates of E. bieneusi in bovine animals were likely caused by discrepancies in animal species, host immune status, geographic regions, sampling sizes as well as management practices.
Notably, the infection of E. bieneusi in yaks was significantly related with age groups (Table 1). In this study, E. bieneusi was found in all age groups, with positive rates of 41.0% (16/39), 11.8% (6/51), 39.5% (16/43) and 15.6% (15/90) for yaks aged <6 months, 6–12 months, 12–24 months and >24 months, respectively, indicating lower positive rate in older yaks (>6 months) compared with younger yaks (<6 months). Similar results have also been reported in dairy cattle in Brazil and the Czech Republic [3, 13], indicating immunity to E. bieneusi in bovine animals likely increasing with the age. However, contrary results were found in a cross-sectional survey on the positive rates of E. bieneusi in dairy cattle on large farms across multiple states in the United States. Similarly, a longitudinal study of E. bieneusi on a dairy farm in Maryland, USA showed a higher positive rate of E. bieneusi in dairy cattle aged 7–24 months compared with animals aged under 6 months [6, 7, 37, 40].
Sequence analysis based on the ITS gene locus of 53 isolates in yaks found seven known genotypes and five novel genotypes, which could enrich our understanding of the genetic diversity of E. bieneusi in bovine animals. BEB4 was first found in cattle [44], and then reported in humans [34], pigs [60] and nonhuman primates [14]. Genotype J was not only reported in dairy cattle [17], but was also found in humans [60], nonhuman primates [56], donkeys [57], zebras, bears and meerkats [18], chickens [33] and wastewater treatment plants [53], indicating its wide host range and zoonotic potential. EbpC is also a genotype with zoonotic potential, and it has been reported in humans [42], nonhuman primates [54] and quite a few animals, such as pigs [8], sheep and cattle [12], dogs [15] and horses [32]. LQ10 was previously reported in Marmota baibacina (ON165748) in China, and the occurrence of this subtype in the present study enriched its host range. Genotype LW1 was first reported in lake water in China [54], and then identified in humans [49], swine [31], sheep [9] and deer [39], indicating the wide host range and zoonotic potential of this genotype. Meanwhile, genotype PigEBITS5 was first reported in swine in Massachusetts, USA [2], and then in humans [35], dogs [15], house mice [36] and raw wastewater [53], reflecting the broad host range and zoonotic potential of this genotype. Genotype ESH-01 has been reported in wastewater [27] and horses [32], and further studies are needed to explore its host range. Furthermore, five novel genotypes, namely Ganzi1–5, were first detected in the present study, and the host range and zoonotic potential of these genotypes will need to be evaluated in the future. Interestingly, all genotypes were J in yaks from Yajiang, while this genotype was absent in Seda and Litang, reflecting low genotypic diversity and unique genotypic distribution of yaks in Yajiang compared with the other two locations.
Further phylogenetic analysis indicated the occurrence of genotypes from both group 1 and group 2 with zoonotic potential in yaks in the present study, which is consistent with genotypes identified in yaks in Tibet [51, 58], Qinghai [26] and Gansu [29] and a cross-sectional survey across Qinghai, Yunnan and Tibet in China [59], reflecting zoonotic potential of these animals in the transmission of E. bieneusi. Two genotypes (CHN11 and CHN12) from group 1 and three genotypes (BEB4, I and J) from group 2 were identified in yaks in Qinghai [26]. Two genotypes (CHN14 and D) from group 1 and eight genotypes (I, J, BEB4, BEB6, COS-I, NESH5, CHC8 and CHN13) from group 2 were found in yaks in Tibet [51, 58]. WCY1 from group 1 and two genotypes (I and BEB4) from group 2 were reported in yaks in Gansu [29]. Yak1 from group 1 and BEB6 from group 2 were recognised in yaks in the cross-sectional survey across Qinghai, Yunnan and Tibet in China [59]. Compared with previous reports in yaks [26, 29, 51, 58, 59], the present study first identified the occurrence of ESH-01, PigEBITS5, LQ10, LW1, EpbC and three novel genotypes (Ganzi1, Ganzi2 and Ganzi4) from group 1 and novel Ganzi3 from group 2 in yaks, reflecting the possible expansion of genotypes in yaks. Among the five novel genotypes identified in yaks in this study, it is interesting to note that genotype Ganzi5 formed a separate branch in the phylogenetic tree, different from other genotypes with various host ranges within groups 1–11, indicating that Ganzi5 could be adapted to yaks, but more work needs to be done in the future to confirm this. Considering the increasing genotypes with zoonotic potential of E. bieneusi in yaks, interventions should be considered to prevent cross-transmission of E. bieneusi between humans and animals.
Conclusions
In this study, we investigated E. bieneusi infection in yaks in Ganzi Tibetan Autonomous Prefecture, Sichuan Province, with a total positive rate of 23.8%, and significant differences were identified in the positive rates among location and age groups. A total of 12 genotypes, including seven known and five novel genotypes, were identified in yaks. Of these identified genotypes, eight and three genotypes were from the group 1 and group 2, respectively indicating potential importance and zoonotic potential of E. bieneusi in these yaks. These findings provide fundamental data for understanding transmission of E. bieneusi in yaks as well as other bovine animals.
Conflicts of interest
Authors declare that they have no conflict of interest.
Funding
This project was supported by the Scientific Research Foundation of the Northwest A&F University (2452021058; 2452022158) and National Natural Science Foundation of Sichuan Province (2023NSFSC0179).
References
- Breton J, Bart-Delabesse E, Biligui S, Carbone A, Seiller X, Okome-Nkoumou M, Nzamba C, Kombila M, Accoceberry I, Thellier M. 2007. New highly divergent rRNA sequence among biodiverse genotypes of Enterocytozoon bieneusi strains isolated from humans in Gabon and Cameroon. Journal of Clinical Microbiology, 45(8), 2580–2589. [CrossRef] [PubMed] [Google Scholar]
- Buckholt MA, Lee JH, Tzipori S. 2002. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Applied and Environmental Microbiology, 68(5), 2595–2599. [CrossRef] [PubMed] [Google Scholar]
- da Silva Fiuza VR, Lopes CW, de Oliveira FC, Santin M. 2016. New findings of Enterocytozoon bieneusi in beef and dairy cattle in Brazil. Veterinary Parasitology, 216, 46–51. [CrossRef] [PubMed] [Google Scholar]
- Didier ES, Weiss LM. 2006. Microsporidiosis: current status. Current Opinion in Infectious Diseases, 19(5), 485–492. [CrossRef] [PubMed] [Google Scholar]
- Du M, Zhang C, Zhao Z, Gu D, Li G, Yi P. 2020. Infection status investigation and species identification of Cryptosporidium in resident yak calves in Datong County of Qinghai Province. Animal Husbandry and Feed Science, 41(3), 106–110. [Google Scholar]
- Fayer R, Santin M, Macarisin D. 2012. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitology Research, 111(3), 1349–1355. [CrossRef] [PubMed] [Google Scholar]
- Fayer R, Santín M, Trout JM. 2007. Enterocytozoon bieneusi in mature dairy cattle on farms in the eastern United States. Parasitology Research, 102(1), 15–20. [CrossRef] [PubMed] [Google Scholar]
- Feng Y, Li N, Dearen T, Lobo ML, Matos O, Cama V, Xiao L. 2011. Development of a multilocus sequence typing tool for high-resolution genotyping of Enterocytozoon bieneusi. Applied and Environmental Microbiology, 77(14), 4822–4828. [PubMed] [Google Scholar]
- Fiuza V, Lopes CWG, Cosendey RIJ, de Oliveira FCR, Fayer R, Santín M. 2016. Zoonotic Enterocytozoon bieneusi genotypes found in Brazilian sheep. Research in Veterinary Science, 107, 196–201. [CrossRef] [PubMed] [Google Scholar]
- Han B, Pan G, Weiss LM. 2021. Microsporidiosis in humans. Clinical Microbiology Reviews, 34(4), e0001020. [CrossRef] [PubMed] [Google Scholar]
- Han B, Weiss LM. 2017. Microsporidia: obligate intracellular pathogens within the Fungal Kingdom. Microbiology Spectrum, 5(2). https://doi.org/10.1128/microbiolspec.FUNK-0018-2016. [PubMed] [Google Scholar]
- Jiang Y, Tao W, Wan Q, Li Q, Yang Y, Lin Y, Zhang S, Li W. 2015. Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep and cattle in Northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Applied and Environmental Microbiology, 81(10), 3326–3335. [Google Scholar]
- Juránková J, Kamler M, Kovařčík K, Koudela B. 2013. Enterocytozoon bieneusi in Bovine Viral Diarrhea Virus (BVDV) infected and noninfected cattle herds. Research in Veterinary Science, 94(1), 100–104. [CrossRef] [PubMed] [Google Scholar]
- Karim MR, Dong H, Li T, Yu F, Li D, Zhang L, Li J, Wang R, Li S, Li X, Rume FI, Ning C. 2015. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS One, 10(2), e0117991. [CrossRef] [PubMed] [Google Scholar]
- Karim MR, Dong H, Yu F, Jian F, Zhang L, Wang R, Zhang S, Rume FI, Ning C, Xiao L. 2014. Genetic diversity in Enterocytozoon bieneusi isolates from dogs and cats in China: host specificity and public health implications. Journal of Clinical Microbiology, 52(9), 3297–3302. [Google Scholar]
- Köster PC, Lapuente J, Pizarro A, Prieto-Pérez L, Pérez-Tanoira R, Dashti A, Bailo B, Muadica AS, González-Barrio D, Calero-Bernal R, Ponce-Gordo F, Carmena D. 2021. Presence and genetic diversity of enteric protists in captive and semi-captive non-human primates in Côte d’Ivoire, Sierra Leone, and Peru. International Journal for Parasitology: Parasites and Wildlife, 17, 26–34. [Google Scholar]
- Li J, Luo N, Wang C, Qi M, Cao J, Cui Z, Huang J, Wang R, Zhang L. 2016. Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia. China. Parasites & Vectors, 9, 142. [CrossRef] [Google Scholar]
- Li J, Qi M, Chang Y, Wang R, Li T, Dong H, Zhang L. 2015. Molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in captive wildlife at Zhengzhou Zoo, China. Journal of Eukaryotic Microbiology, 62(6), 833–839. [CrossRef] [Google Scholar]
- Li S, Wang P, Zhu XQ, Zou Y, Chen XQ. 2022. Prevalence and genotypes/subtypes of Enterocytozoon bieneusi and Blastocystis sp. in different breeds of cattle in Jiangxi Province, southeastern China. Infection, Genetics and Evolution, 98, 105216. [CrossRef] [PubMed] [Google Scholar]
- Li W, Feng Y, Santin M. 2019. Host specificity of Enterocytozoon bieneusi and public health implications. Trends in Parasitology, 35(6), 436–451. [CrossRef] [PubMed] [Google Scholar]
- Li W, Feng Y, Zhang L, Xiao L. 2019. Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infection, Genetics and Evolution, 75, 104033. [CrossRef] [PubMed] [Google Scholar]
- Liu X. 2022. A study on the prevention and control of yak hydatidosis in high altitude areas of Qinghai Province. Farmers Consultant, 731(12), 111–113. [Google Scholar]
- Liu X, Tang L, Li W, Li C, Gu Y. 2022. Prevalence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi from large-scale cattle farms in Anhui Province, China. Journal of Veterinary Medical Science, 84(1), 40–47. [CrossRef] [PubMed] [Google Scholar]
- Lobo ML, Xiao L, Antunes F, Matos O. 2012. Microsporidia as emerging pathogens and the implication for public health: a 10-year study on HIV-positive and -negative patients. International Journal for Parasitology, 42(2), 197–205. [CrossRef] [PubMed] [Google Scholar]
- Lobo ML, Xiao L, Cama V, Stevens T, Antunes F, Matos O. 2006. Genotypes of Enterocytozoon bieneusi in mammals in Portugal. Journal of Eukaryotic Microbiology, 53(Suppl 1), S61–S64. [Google Scholar]
- Ma J, Cai J, Ma J, Feng Y, Xiao L. 2015. Enterocytozoon bieneusi genotypes in yaks (Bos grunniens) and their public health potential. Journal of Eukaryotic Microbiology, 62(1), 21–25. [CrossRef] [Google Scholar]
- Ma J, Feng Y, Hu Y, Villegas EN, Xiao L. 2016. Human infective potential of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in urban wastewater treatment plant effluents. Journal of Water and Health, 14(3), 411–423. [CrossRef] [PubMed] [Google Scholar]
- Ma J, Li P, Zhao X, Xu H, Wu W, Wang Y, Guo Y, Wang L, Feng Y, Xiao L. 2015. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Veterinary Parasitology, 207(3–4), 220–227. [CrossRef] [PubMed] [Google Scholar]
- Ma JG, Zhang NZ, Hou JL, Zou Y, Hu GX, Zhu XQ, Zhou DH. 2017. Detection of Enterocytozoon bieneusi in white yaks in Gansu Province, China. BioMed Research International, 2017, 5790181. [PubMed] [Google Scholar]
- Mathis A, Weber R, Deplazes P. 2005. Zoonotic potential of the microsporidia. Clinical Microbiology Reviews, 18(3), 423–445. [CrossRef] [PubMed] [Google Scholar]
- Němejc K, Sak B, Květoňová D, Hanzal V, Janiszewski P, Forejtek P, Rajský D, Kotková M, Ravaszová P, McEvoy J, Kváč M. 2014. Prevalence and diversity of Encephalitozoon spp. and Enterocytozoon bieneusi in wild boars (Sus scrofa) in Central Europe. Parasitology Research, 113(2), 761–767. [CrossRef] [PubMed] [Google Scholar]
- Qi M, Wang R, Wang H, Jian F, Li J, Zhao J, Dong H, Zhu H, Ning C, Zhang L. 2016. Enterocytozoon bieneusi genotypes in grazing horses in China and their zoonotic transmission potential. Journal of Eukaryotic Microbiology, 63(5), 591–597. [CrossRef] [Google Scholar]
- Reetz J, Rinder H, Thomschke A, Manke H, Schwebs M, Bruderek A. 2002. First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens). International Journal for Parasitology, 32(7), 785–787. [CrossRef] [PubMed] [Google Scholar]
- Sak B, Brady D, Pelikánová M, Květoňová D, Rost M, Kostka M, Tolarová V, Hůzová Z, Kváč M. 2011. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. Journal of Clinical Microbiology, 49(3), 1064–1070. [Google Scholar]
- Sak B, Kváč M, Kučerová Z, Květoňová D, Saková K. 2011. Latent microsporidial infection in immunocompetent individuals – a longitudinal study. PLoS Neglected Tropical Diseases, 5(5), e1162. [CrossRef] [PubMed] [Google Scholar]
- Sak B, Kváč M, Květoňová D, Albrecht T, Piálek J. 2011. The first report on natural Enterocytozoon bieneusi and Encephalitozoon spp. infections in wild East-European House Mice (Mus musculus musculus) and West-European House Mice (M. m. domesticus) in a hybrid zone across the Czech Republic-Germany border. Veterinary Parasitology, 178(3–4), 246–250. [CrossRef] [PubMed] [Google Scholar]
- Santín M, Fayer R. 2009. A longitudinal study of Enterocytozoon bieneusi in dairy cattle. Parasitology Research, 105(1), 141–144. [CrossRef] [PubMed] [Google Scholar]
- Santín M, Fayer R. 2011. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Research in Veterinary Science, 90(3), 363–371. [CrossRef] [PubMed] [Google Scholar]
- Santin M, Fayer R. 2015. Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. Journal of Eukaryotic Microbiology, 62(1), 34–43. [CrossRef] [Google Scholar]
- Santín M, Trout JM, Fayer R. 2005. Enterocytozoon bieneusi genotypes in dairy cattle in the eastern United States. Parasitology Research, 97(6), 535–538. [CrossRef] [PubMed] [Google Scholar]
- Stentiford GD, Becnel J, Weiss LM, Keeling PJ, Didier ES, Williams BP, Bjornson S, Kent ML, Freeman MA, Brown MJF, Troemel ER, Roesel K, Sokolova Y, Snowden KF, Solter L. 2016. Microsporidia – emergent pathogens in the global food chain. Trends in Parasitology, 32(4), 336–348. [CrossRef] [PubMed] [Google Scholar]
- Sulaiman IM, Bern C, Gilman R, Cama V, Kawai V, Vargas D, Ticona E, Vivar A, Xiao L. 2003. A molecular biologic study of Enterocytozoon bieneusi in HIV-infected patients in Lima, Peru. Journal of Eukaryotic Microbiology, 50(Suppl), 591–596. [CrossRef] [PubMed] [Google Scholar]
- Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW, Xiao L. 2003. Molecular characterization of microsporidia indicates that wild mammals Harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Applied and Environmental Microbiology, 69(8), 4495–4501. [CrossRef] [PubMed] [Google Scholar]
- Sulaiman IM, Fayer R, Yang C, Santin M, Matos O, Xiao L. 2004. Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitology Research, 92(4), 328–334. [CrossRef] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725–2729. [CrossRef] [PubMed] [Google Scholar]
- Tang W, Nie F, Zhang Y, Jia Z, Yi F. 2020. Investigation on Toxoplasma gondii infection in Tianzhu white yaks. China Herbivore Science, 40(3), 91–92. [Google Scholar]
- Tao WF, Ni HB, Du HF, Jiang J, Li J, Qiu HY, Li Y, Zhang XX. 2020. Molecular detection of Cryptosporidium and Enterocytozoon bieneusi in dairy calves and sika deer in four provinces in Northern China. Parasitology Research, 119(1), 105–114. [CrossRef] [PubMed] [Google Scholar]
- Thellier M, Breton J. 2008. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite, 15(3), 349–358. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Wang L, Xiao L, Duan L, Ye J, Guo Y, Guo M, Liu L, Feng Y. 2013. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Neglected Tropical Diseases, 7(9), e2437. [CrossRef] [PubMed] [Google Scholar]
- Wang XT, Wang RJ, Ren GJ, Yu ZQ, Zhang LX, Zhang SY, Lu H, Peng XQ, Zhao GH. 2016. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi province, northwestern China. Parasitology Research, 115(3), 1355–1361. [CrossRef] [PubMed] [Google Scholar]
- Wu Y, Chang Y, Zhang X, Chen Y, Li D, Wang L, Zheng S, Wang R, Zhang S, Jian F, Ning C, Li J, Zhang L. 2019. Molecular characterization and distribution of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi from yaks in Tibet, China. BMC Veterinary Research, 15(1), 417. [CrossRef] [PubMed] [Google Scholar]
- Xiao L. 2015. Biology of foodborne parasites. Florida: CRC Press. [CrossRef] [Google Scholar]
- Ye J, Ji Y, Xu J, Ma K, Yang X. 2017. Zoonotic Enterocytozoon bieneusi in raw wastewater in Zhengzhou, China. Folia Parasitologica, 64, 002. [Google Scholar]
- Ye J, Xiao L, Ma J, Guo M, Liu L, Feng Y. 2012. Anthroponotic enteric parasites in monkeys in public park, China. Emerging Infectious Diseases, 18(10), 1640–1643. [CrossRef] [PubMed] [Google Scholar]
- Yin RH, Bai WL, Wang JM, Wu CD, Dou QL, Yin RL, He JB, Luo GB. 2009. Development of an assay for rapid identification of meat from yak and cattle using polymerase chain reaction technique. Meat Science, 83(1), 38–44. [CrossRef] [PubMed] [Google Scholar]
- Yu F, Wu Y, Li T, Cao J, Wang J, Hu S, Zhu H, Zhang S, Wang R, Ning C, Zhang L. 2017. High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Veterinary Research, 13(1), 158. [CrossRef] [PubMed] [Google Scholar]
- Yue DM, Ma JG, Li FC, Hou JL, Zheng WB, Zhao Q, Zhang XX, Zhu XQ. 2017. Occurrence of Enterocytozoon bieneusi in donkeys (Equus asinus) in China: a public health concern. Frontiers in Microbiology, 8, 565. [PubMed] [Google Scholar]
- Zhang Q, Cai J, Li P, Wang L, Guo Y, Li C, Lei M, Feng Y, Xiao L. 2018. Enterocytozoon bieneusi genotypes in Tibetan sheep and yaks. Parasitology Research, 117(3), 721–727. [CrossRef] [PubMed] [Google Scholar]
- Zhang Q, Zhang Z, Ai S, Wang X, Zhang R, Duan Z. 2019. Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis from animal sources in the Qinghai-Tibetan Plateau Area (QTPA) in China. Comparative Immunology, Microbiology and Infectious Diseases, 67, 101346. [CrossRef] [PubMed] [Google Scholar]
- Zhang X, Wang Z, Su Y, Liang X, Sun X, Peng S, Lu H, Jiang N, Yin J, Xiang M, Chen Q. 2011. Identification and genotyping of Enterocytozoon bieneusi in China. Journal of Clinical Microbiology, 49(5), 2006–2008. [Google Scholar]
- Zhang Y, Koehler AV, Wang T, Gasser RB. 2021. Enterocytozoon bieneusi of animals-with an “Australian twist”. Advances in Parasitology, 111, 1–73. [CrossRef] [PubMed] [Google Scholar]
- Zhang Y, Koehler AV, Wang T, Robertson GJ, Bradbury RS, Gasser RB. 2018. Enterocytozoon bieneusi genotypes in people with gastrointestinal disorders in Queensland and Western Australia. Infection, Genetics and Evolution, 65, 293–299. [CrossRef] [PubMed] [Google Scholar]
- Zhao W, Zhang W, Yang F, Zhang L, Wang R, Cao J, Shen Y, Liu A. 2015. Enterocytozoon bieneusi in dairy cattle in the Northeast of China: genetic diversity of ITS gene and evaluation of zoonotic transmission potential. Journal of Eukaryotic Microbiology, 62(4), 553–560. [CrossRef] [Google Scholar]
Cite this article as: Yang X, Fan Y-Y, Yang D-J, Huang S, Wang J-W, Chen X, Zhang M, Liu Y-W, Li Q, Song J-K & Zhao G-H. 2023. High genotype diversity and zoonotic potential of Enterocytozoon bieneusi in yaks (Bos grunniens) from Ganzi Tibetan Autonomous Prefecture, Sichuan Province. Parasite 30, 39.
All Tables
Occurrence, genotypes and factors associated with E. bieneusi infection in yaks from Ganzi Tibetan Autonomous Prefecture.
All Figures
 |
Figure 1 Geographical distribution of sampling sites in Ganzi Tibetan Autonomous Prefecture, Sichuan Province, China. |
| In the text | |
 |
Figure 2 Phylogenetic relationships of E. bieneusi genotypes in this study, with reference sequences downloaded from GenBank based on the sequence analysis of the ITS locus using Maximum Likelihood analysis with the General Time Reversible model. Red and blue filled circles before the bold sample names represent novel and known genotypes identified in the present study, respectively. Bootstrap values (> 50) are indicated above the nodes. Scale bar indicates 0.1 nucleotide substitutions/site. Genotype SW3 from stormwater (KF591679.1) is used as the outgroup. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


