| Issue |
Parasite
Volume 24, 2017
|
|
|---|---|---|
| Article Number | 30 | |
| Number of page(s) | 8 | |
| DOI | https://doi.org/10.1051/parasite/2017025 | |
| Published online | 02 August 2017 | |
Research Article
Sarcocystis spp. in domestic sheep in Kunming City, China: prevalence, morphology, and molecular characteristics
Sarcocystis spp. chez les moutons domestiques à Kunming, en Chine : prévalence, morphologie et caractéristiques moléculaires
1
School of Biological Sciences, Yunnan University, Kunming
650091, PR China
2
Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin, Nay Pyi Taw
05282, Myanmar
3
Department of Biology, Wake Forest University, Winston-Salem, NC
27106, USA
* Corresponding author: jjhu@ynu.edu.cn
Received:
6
May
2017
Accepted:
9
June
2017
Sheep (Ovis aries) are intermediate hosts for at least six named species of Sarcocystis: S. tenella, S. arieticanis, S. gigantea, S. medusiformis, S. mihoensis, and S. microps. Here, only two species, S. tenella and S. arieticanis, were found in 79 of 86 sheep (91.9%) in Kunming, China, based on their morphological characteristics. Four genetic markers, i.e., 18S rRNA gene, 28S rRNA gene, mitochondrial cox1 gene, and ITS-1 region, were sequenced and characterized for the two species of Sarcocystis. Sequences of the three former markers for S. tenella shared high identities with those of S. capracanis in goats, i.e., 99.0%, 98.3%, and 93.6%, respectively; the same three marker sequences of S. arieticanis shared high identities with those of S. hircicanis in goats, i.e., 98.5%, 96.5%, and 92.5%, respectively. No sequences in GenBank were found to significantly resemble the ITS-1 regions of S. tenella and S. arieticanis. Identities of the four genetic markers for S. tenella and S. arieticanis were 96.3%, 95.4%, 82.5%, and 66.2%, respectively.
Résumé
Les moutons (Ovis aries) sont des hôtes intermédiaires pour au moins 6 espèces nommées de Sarcocystis : S. tenella, S. arieticanis, S. gigantea, S. medusiformis, S. mihoensis et S. microps. Ici, seules deux espèces, S. tenella et S. arieticanis ont été trouvées dans 79 (91.9 %) de 86 moutons à Kunming, en Chine, en fonction de leurs caractéristiques morphologiques. Quatre marqueurs génétiques, le gène de l’ARNr 18S, le gène de l’ARNr 28S, le gène mitochondrial cox1 et la région de l’ITS-1 ont été séquencés et caractérisés pour les deux espèces de Sarcocystis. Les séquences des trois premiers marqueurs de S. tenella partageaient des similarités élevées avec celles de S. capracanis chez les chèvres, soit 99.0 %, 98.3 % et 93.6 %, respectivement; les trois mêmes marqueurs de S. arieticanis partageaient des identités élevées avec ceux de S. hircicanis chez les chèvres, à savoir 98.5 %, 96.5 % et 92.5 %, respectivement. Aucune séquence de GenBank ne ressemble significativement aux régions ITS-1 de S. tenella et S. arieticanis. Les identités des quatre marqueurs génétiques de S. tenella et S. arieticanis étaient respectivement de 96.3 %, 95.4 %, 82.5 % et 66.2 %.
Key words: Sheep / Sarcocystis / Prevalence / Morphology / Molecular characteristics / China
© J.-J. Hu et al., published by EDP Sciences, 2017
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Sarcocystis spp. are cyst-forming intracellular protozoan parasites with an obligate two-host life cycle between predators as definitive hosts and prey animals as intermediate hosts. Sheep (Ovis aries) are intermediate hosts for at least six species, i.e., S. tenella, S. arieticanis, S. gigantea, S. medusiformis, S. mihoensis, and S. microps, which are morphologically differentiated based on their sarcocyst wall ultrastructure. Sarcocystis tenella and S. arieticanis produce microscopic sarcocysts transmitted by canids, while S. gigantea and S. medusiformis produce macroscopic cysts transmitted by felids [2]. The remaining two species, S. mihoensis and S. microps transmitted by canids, are unusual or rare species of Sarcocystis; S. mihoensis, reported only from Japan, produces macroscopic sarcocysts [17]; however, S. microps, reported only once from China, produces microscopic sarcocysts [18]. Natural infections by Sarcocystis spp. in domestic sheep have been investigated in various countries throughout the world, with prevalence ranging from 9.0 to 100% depending on the detection methodology [2]. However, the prevalence of Sarcocystis spp. in domestic sheep in China is largely unknown.
The ultrastructure of sarcocysts is traditionally a reliable character for clarifying different Sarcocystis species in a given intermediate host. However, with more morphologically similar sarcocysts described from different, but closely related intermediate hosts, confusions or disputes have emerged concerning the relationships of these Sarcocystis species. For example, Sarcocystis spp. in cattle and water buffalo have been regarded as separate species based on host specificity; however, sarcocysts in bobcats (Felis rufus), domestic cats (F. catus), Florida panthers and cougars (F. concolor), and cheetahs (Acinonyx jubatus) were all identified as S. felis, on the basis of the morphological similarities [2]. It is therefore an urgent need, even a must, to delineate or reassess descriptions of extant or new species of Sarcocystis in different hosts, using different markers for clarifying their relationships. However, only limited molecular sequences for Sarcocystis spp. in sheep are presently provided in GenBank.
Therefore, the aims of the present study were (i) to investigate the prevalence of Sarcocystis spp. in domestic sheep in China based on the morphological characteristics of the sarcocysts, and (ii) to characterize these species using the 18S rRNA gene (18S rRNA), 28S rRNA gene (28S rRNA), mitochondrial cox1 gene (cox1), and ITS-1 (ITS-1) region for clarifying their descriptions.
Materials and methods
Morphological observation of sarcocysts
In total, tissues from 86 sheep were examined from an abattoir in Kunming City in China from March to November 2015. From each animal, fresh tissue samples from the esophagus, diaphragm, skeletal muscles, tongue, and heart were examined for sarcocysts. In the laboratory, 0.5 mm pieces of muscle from each collected sample were pressed and squeezed between two glass slides to inspect sarcocysts using stereomicroscopy. Sarcocysts were isolated from muscular fibers using dissecting needles and processed for light microscopy (LM), transmission electron microscopy (TEM), and DNA analysis.
For TEM, sarcocysts were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4) at 4 °C and post-fixed in 1% osmium tetroxide in the same buffer, then dehydrated in graded alcohols and embedded in epon-araldite mixture. Ultrathin sections were stained with uranyl acetate and lead citrate and then examined using a JEM100-CX transmission electron microscope at 80 kV. For DNA isolation, individual cysts were stored in sterile water at −20 °C prior to processing.
Molecular characterization
Two individual sarcocysts of each of the Sarcocystis species from sheep were subjected to genomic DNA extraction using the phenol/chloroform method after 0.01% proteinase K and 0.25% trypsin digestion. 18S rRNA was amplified with primer pairs, S1/B [4, 11]; 28S rRNA was amplified with primer sets KL1/KL3, KL4/KL5b, and KL6/KL2 [12]; and mitochondrial cox1 was amplified with primer pairs SF1/SR9 [6, 7]; ITS-1 was amplified with primer pairs SU1F/5.8SR2 [8]. The polymerase chain reaction (PCR) products were purified, cloned, sequenced, and analyzed using the method detailed in a previous paper [9].
Results
Prevalence of natural infections
Sarcocysts were found in 79 of 86 sheep (91.9%). Two morphologically distinct sarcocysts (S. tenella with a thick cyst wall and S. arieticanis with a thin cyst wall) were observed by LM observation (Figs. 1A, 2A). Sarcocysts of S. tenella were found in 73 sheep (84.9%), and were more common than those of S. arieticanis, found in 46 sheep (53.5%). The distribution of the two parasites in different organs is shown in Table 1.
 |
Figure 1. Morphological characteristics of Sarcocystis tenella sarcocysts from sheep muscle. (A) Light micrograph of a sarcocyst (unstained); note the sarcocyst wall with numerous palisade-like protrusions (pp). Scale bar = 20 μm. (B) Longitudinal section of sarcocyst wall by transmission electron microscopy (TEM). The sarcocyst wall had palisade-like protrusions (pp), characterized by the apex which contained dense plaques (arrow); a layer of ground substances (gs) located beneath the primary sarcocyst wall surrounded bradyzoites (bz). Scale bar = 1 μm. (C) Cross-section of a sarcocyst by TEM; note the dense plaques (arrow), ground substance (gs), bradyzoites (bz), and host cell (hc). Scale bar = 1 μm. |
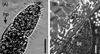 |
Figure 2. Morphological characteristics of Sarcocystis arieticanis sarcocysts from sheep muscle. (A) Light micrograph of a sarcocyst (unstained); note the sarcocyst wall with numerous hair-like protrusions (hp). Scale bar = 20 μm. (B) Transmission electron micrograph of the cyst wall of a sarcocyst; note host cell (hc), bradyzoites (bz), and hair-like protrusions (arrow). Scale bar = 1 μm. |
Prevalence of Sarcocystis spp. in different muscular tissues of sheep (n = 86) in Kunming, China.
Light and electron microscopy observations of sarcocysts
Using LM, sarcocysts of S. tenella were observed to be microscopic, measuring 650–1,350 × 65–130 μm (n = 30) in size. The sarcocyst wall had numerous, 3.0–5.8 μm (n = 30) long, palisade-like protrusions (Fig. 1A). The sarcocysts were septate and their interior compartments were filled with bradyzoites measuring 11.0–14.0 × 3.5–5.0 μm (n = 30) in size. By TEM, the sarcocysts had numerous villous or palisade-like protrusions, were 3.1–4.6 × 1.2–1.7 μm (n = 10) in size, and were characterized by the apex that contained dense plaques; microtubules were absent (Figs. 1B, 1C). A layer of ground substances measuring 0.4–0.6 μm in thickness was located immediately beneath the primary sarcocyst wall.
Sarcocysts of S. arieticanis were microscopic, measuring 137–1,272 × 75–103 μm (n = 30) in size (Fig. 2A) by LM. The sarcocyst wall had numerous, 4.7–6.0 μm (n = 30) in length, hair-like protrusions. The cysts were septate and their interior compartments were filled with bradyzoites measuring 9.5–13.6 × 4.0–5.0 μm (n = 30) in size. The ultrastructure of the sarcocysts showed the presence of irregularly folded, but non-branched, hirsute or bone-like protrusions (Fig. 2B). A layer of ground substances measuring 0.3–0.5 μm in thickness was located immediately beneath the primary sarcocyst wall.
Molecular characterization of the 18S rRNA
The two 18S rRNA nucleotide sequences, each from an individual sarcocyst of S. tenella, were 1,832 bp in length, and completely identical; as a result, only one sequence (MF039329) was submitted to GenBank. The most similar sequences in GenBank were those of S. tenella (KC209734 and KC209737) from sheep (99.8–99.9% identity, on average 99.9% identity), followed by S. tenella (KP263752–KP263759) from chamois (Rupicapra rupicapra) (99.7–99.9% identity, on average 99.8% identity), S. capracanis (L76472, KU820982, and KU820983) from goats (Capra hircus) (99.0–99.1% identity, on average 99.0% identity), S. alces (KF831273 and KF831274) from moose (Alces alces) (97.6–97.8% identity, on average 97.6% identity), S. heydorni (KX057996 and KX057997) from cattle (Bos taurus) (97.5–97.6% identity, on average 97.5% identity), S. tarandivulpes (EF056012) from reindeer (Rangifer tarandus) (97.3% identity), and S. cruzi (JX679468) from cattle (97.0% identity).
The two 18S rRNA sequences (MF039330 and MF039331), each obtained from an individual sarcocyst of S. arieticanis, were 1,836 bp in length, and shared 99.9% identity. The differences included three nucleotide substitutions. Identity of the new 18S RNA sequences between S. arieticanis and S. tenella was 96.1–96.2% and 96.2% on average. The most similar sequences in GenBank were those of S. hircicanis (KU820984 and KU820985) from goats (98.5–98.6% identity, on average 98.5% identity), followed by S. arieticanis (L24382) from sheep (98.3–98.5% identity, on average 98.4% identity), S. cruzi (KT901167, JX679467, and JX679468) from cattle (96.5–96.8% identity, on average 96.6% identity), S. hjorti (JX679468) in moose (96.3–96.4% identity, on average 96.3% identity), S. levinei (KU247914–KU247922) from water buffalos (Bubalus bubalis) (96.4–96.5% identity, on average 96.5% identity), S. pilosa (KU753891–KU753893) from sika deer (Cervus nippon) (96.3–96.4% identity, on average 96.3% identity), and S. tenella (KC209737) from sheep (96.2–96.3% identity, on average 96.3% identity).
Molecular characterization of the 28S rRNA
The two 28S rRNA nucleotide sequences (MF039325 and MF039326), each obtained from an individual sarcocyst of S. tenella, were 3,459 bp in length, and shared 99.8% identity. The differences included six nucleotide substitutions. The most similar sequence in GenBank was that of S. tenella (AF076899) (98.9–99.1% identity, on average 99.0% identity), followed by S. capracanis (AF012885, KU820978, and KU820979) (98.2–98.4% identity, on average 98.3% identity), S. cruzi (AF076903) (95.6–95.8% identity, on average 95.7% identity), S. arieticanis (AF076904) (95.4–95.5% identity, on average 95.4% identity), and S. hircicanis (KU820980 and KU820981) (94.3–95.0% identity, on average 94.6% identity).
The two 28S rRNA sequences (MF039327 and MF039328), each obtained from an individual sarcocyst of S. arieticanis, were 3,482 bp and 3,506 bp in length, respectively, and shared 97.8% identity. The differences included 42 nucleotide substitutions and 23 nucleotide deletions. Identity of the new 28S rRNA sequences between S. arieticanis and S. tenella was 95.3–96.4%, and 95.9% on average. The most similar 28S rRNA sequences in GenBank were those of S. arieticanis (AF076904) (97.6–99.1% identity, on average 98.3% identity), followed by S. hircicanis (KU820980 and KU820981) (96.4–96.7% identity, on average 96.5% identity), S. tenella (AF076899) (96.0–96.8% identity, on average 96.4% identity), S. capracanis (AF012885, KU820978, and KU820979) (95.7–96.5% identity, on average 96.1% identity), and S. cruzi (AF076903) (95.0–95.1% identity, on average 95.0% identity).
Molecular characterization of the mitochondrial cox1
The two mitochondrial cox1 nucleotide sequences (MF039322 and MF039323), each from an individual sarcocyst of S. tenella, were 1,038 bp in length, and shared 98.3% identity. The differences included 15 nucleotide substitutions. They shared the highest identities with those of S. tenella (KC209723–KC209732) from sheep (96.9–97.5%, on average 97.2%) and S. tenella (KP263744–KP2637451) from chamois (96.9–97.5% identity, on average 97.2% identity), followed by S. capracanis (KU920974, KU820977) (93.6% identity), and S. heydorni (KX057994 and KX057995) (90.1–90.3% identity, on average 90.2% identity).
The two mitochondrial cox1 nucleotide sequences, each obtained from an individual sarcocyst of S. arieticanis, were 1,038 bp in length, and shared 100% identity; accordingly, only one sequence (MF039324) was submitted to GenBank. The cox1 nucleotide sequence identity between those for S. arieticanis and S. tenella was 82.2–82.6%, and 82.4% on average. The most similar cox1 sequences in GenBank were those of S. hircicanis (KU820975 and KU8209756) (92.4–92.7% identity, on average 92.5% identity), followed by S. grueneri (KC209615–KC209624) from reindeer (83.7–84.5% identity, on average 84.2% identity), S. capreolicanis (KY018939–KY018944) from roe deer (Capreolus capreolus) (82.6–82.9% identity, on average 82.8% identity), S. capracanis (KU820974 and KU820977) (82.5–82.8%, on average 82.6%), and S. tenella (KC209725 and KC209731) (82.4–82.6% identity, on average 82.5% identity).
Molecular characterization of the ITS-1
The two ITS-1 nucleotide sequences (MF039318 and MF039319), each from a sarcocyst of S. tenella, were 784 bp and 787 bp in length, respectively; the identity between them was 96.7%, and the differences included 23 nucleotide substitutions and three nucleotide deletions. The two ITS-1 nucleotide sequences (MF039320 and MF039321), each from a sarcocyst of S. arieticanis, were 784 bp and 786 bp in length, respectively; the identity between them was 97.0%, and the differences included 20 nucleotide substitutions and four nucleotide deletions. Blast search only using the ITS-1 region, of approximately 525 bp for S. tenella and 520 bp for S. arieticanis, revealed that no sequences shared significant similarities with them. The identity of the new ITS-1 nucleotide sequences for the two species of Sarcocystis in sheep was 65.7–69.7%, and a mean of 66.2%.
The phylogenetic analyses based on either the 18S rRNA, 28S rRNA, or cox1 sequences all clustered the new sequences of S. tenella and S. arieticanis within a clade comprising species with canines as the known, or presumed, definitive hosts (Figs. 3–5).
 |
Figure 3. Phylogenetic tree for selected members of the Sarcocystidae based on 18S rRNA sequences and inferred by the maximum parsimony (MP) method using the Tree-Bisection-Regrafting (TBR) algorithm. The analysis involved 52 nucleotide sequences (GenBank accession numbers behind the taxon names), and a total of 2,014 positions in the final dataset. The values between the branches represent percent bootstrap value per 1,000 replicates, and the values below 50% are not shown. The three new sequences of Sarcocystis tenella (MF039329) and S. arieticanis (MF039330 and MF039331) have the taxon name in boldface. |
 |
Figure 4. Phylogenetic tree for selected members of the Sarcocystidae based on 28S rRNA sequences and inferred by the maximum parsimony (MP) method using the Tree-Bisection-Regrafting (TBR) algorithm. The analysis involved 20 nucleotide sequences (GenBank accession numbers behind the taxon names), and a total of 4,517 positions in the final dataset. The values between the branches represent percent bootstrap value per 1,000 replicates, and the values below 50% are not shown. The four new sequences of Sarcocystis tenella (MF039325 and MF039326) and S. arieticanis (MF039327 and MF039328) have the taxon name in boldface. |
 |
Figure 5. Phylogenetic tree for selected members of the Sarcocystidae based on cox1 sequences and inferred by the maximum parsimony (MP) method using the Tree-Bisection-Regrafting (TBR) algorithm. The analysis involved 27 nucleotide sequences (GenBank accession numbers behind the taxon names), and a total of 998 positions in the final dataset. The values between the branches represent percent bootstrap value per 1,000 replicates, and the values below 50% are not shown. The three new sequences of Sarcocystis tenella (MF039322 and MF039323) and S. arieticanis (MF039324) have the taxon name in boldface. |
Discussion
Sarcocystis spp. are among the most common parasites in domestic ruminants, and some of them can generate significant economic losses when causing clinical and subclinical disease. Up to now, at least six species of Sarcocystis have been named in sheep; however, only four species, i.e., S. tenella, S. arieticanis, S. gigantea, and S. medusiformis, have been frequently found in different areas, especially in Asia and Latin America [2]. In the present study, sarcocysts were common in sheep (91.9%), but only microscopic sarcocysts for S. tenella and S. arieticanis were found, and both parasites were transmitted by canids, which reflects the significant role played by dogs, rather than cats, in the transmission of these parasites in the area.
In the present study, the sarcocyst wall ultrastructures of S. tenella and S. arieticanis belong to “type 14” and “type 7”, respectively, using the classification by Dubey et al. (2016) [2]. Sarcocystis capracanis in goats is a sibling species of S. tenella, and the two species appear similar by LM. However, under TEM, there are some differences in sarcocyst structure, i.e., the presence (S. tenella) or absence (S. capracanis) of disk-like condensations in the apical of the villar protrusions, and the presence (S. capracanis) or absence (S. tenella) of vesicles at the base of the villar protrusions [9]. Sarcocystis tenella-like sarcocysts have also been described from wild Caprinae, i.e., Sarcocystis sp. in wild sheep (Ovis musimon) [13] and Sarcocystis sp. in chamois [14]. The ultrastructure of S. arieticanis cysts is similar to those of Sarcocystis spp. in different Caprinae. For example, S. arieticanis in wild sheep (O. musimon) [16], S. hircicanis in goats [9], S. hircicanis/arieticanis-like in blue sheep (Pseudios nayaur), Japanese serow (Capricornis crispus), and muskox (Ovibos moschatus) [15], plus S. arieticanis-like in Alpine ibex (Capra ibex) [1].
Thus, the two sarcocyst TEM types (14 and 7) in domestic sheep are common in the tissues of different, but closely related, ruminant animals; however, the relationships between these morphologically similar Sarcocystis spp. are not very clear. It is not easy to complete cross-transmission for Sarcocystis spp. using large experimental animals in the laboratory. Accordingly, characterizing sequences of different genetic markers should be a useful tool to differentiate these species of Sarcocystis or delineate their phylogenetic relationships. For example, Sarcocystis sp. in chamois has been designated as S. tenella based on the similarities of their 18S rRNA and cox1 sequences [10].
In the present study, four genetic markers (18S rRNA, 28S rRNA, cox1, and ITS-1) for Sarcocystis spp. in sheep were sequenced and characterized. Among them, sequences of cox1 for S. arieticanis and ITS-1 for Sarcocystis spp. in sheep were the first records in GenBank. When blasting these sequences in GenBank, sequences of 18S rRNA, 28S rRNA, and cox1 for S. tenella shared high identities with those of S. capracanis, i.e., 99.0%, 98.3%, and 93.6%, respectively; S. hircicanis shared high identities with S. arieticanis, i.e., 98.5%, 96.5%, and 92.5%, respectively. Therefore, mitochondrial cox1 seemed to perform better than 18S rRNA and 28S rRNA for distinguishing S. tenella from S. capracanis, and S. arieticanis from S. hircicanis. The high identity (99.0%) between 18S rRNA sequences for S. tenella and S. capracanis has even led to a debate whether sheep and goat can harbor the same Sarcocystis species [3, 5]. Blast search suggested that no sequences in GenBank had significant similarity with the ITS-1 regions of S. tenella and S. arieticanis. However, when comparing the new sequences of the four genetic markers (18S rRNA, 28S rRNA, cox1, and ITS-1) for S. tenella and S. arieticanis, the sequence identities were 96.3%, 95.4%, 82.5%, and 66.2%, respectively. Thus, the ITS-1 region could be more useful for discriminating closely related Sarcocystis spp. because of its high divergence.
Acknowledgments
The present study was funded by the Natural Science Foundation of China (Grant 31460557) and the Southeast Asia Biodiversity Research Institute, Chinese Academy of Science (Grant Y4zk111B01). We would like to thank Dr. J.P. Dubey for his positive comments regarding the content of the paper and his suggestions dealing with the presentation of the text.
References
- Cornaglia E, Giaccherino AR, Peracino V. 1998. Ultrastructural morphology of sarcosporidiosis in Alpine ibex (Capra ibex). Veterinary Parasitology, 75, 21–32. [CrossRef] [PubMed] [Google Scholar]
- Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R. 2016. Sarcocystosis of animals and humans, 2nd edn. Boca Raton, FL: CRC Press. p. 195–214, 217–234, 243–248, 273–275. [Google Scholar]
- Dubey JP, Rosenthal BM. 2013. Sarcocystis capracanis-associated encephalitis in sheep. Veterinary Parasitology, 197, 407–408. [CrossRef] [PubMed] [Google Scholar]
- Fischer S, Odening K. 1998. Characterization of bovine Sarcocystis species by analysis of their 18S ribosomal DNA sequences. Journal of Parasitology, 84, 50–54. [CrossRef] [Google Scholar]
- Formisano P, Aldridge B, Alony Y, Beekhuis L, Davies E, Del Pozo J, Dunn K, English K, Morrison L, Sargison N, Seguino A, Summers BA, Wilson D, Milne E, Beard PM. 2013. Identification of Sarcocystis capracanis in cerebrospinal fluid from sheep with neurological disease. Veterinary Parasitology, 193, 252–255. [CrossRef] [PubMed] [Google Scholar]
- Gjerde B. 2013. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. International Journal for Parasitology, 43, 579–591. [CrossRef] [PubMed] [Google Scholar]
- Gjerde B. 2014. Sarcocystis species in red deer revisited: with a redescription of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology, 141, 441–452. [CrossRef] [PubMed] [Google Scholar]
- Gjerde B. 2014. Molecular characterization of Sarcocystis rileyi from a common eider (Somateria mollissima) in Norway. Parasitology Research, 113, 3501–3509. [CrossRef] [PubMed] [Google Scholar]
- Hu JJ, Liu TT, Liu Q, Esch GW, Chen JQ, Huang S, Wen T. 2016. Prevalence, morphology, and molecular characteristics of Sarcocystis spp. in domestic goats (Capra hircus) from Kunming, China. Parasitology Research, 115, 3973–3981. [CrossRef] [PubMed] [Google Scholar]
- Kolenda R, Schierack P, Zieba F, Zwijacz-Kozica T, Bednarski M. 2015. First molecular characterization of Sarcocystis tenella in Tatra chamois (Rupicapra rupicapra tatrica) in Poland. Parasitology Research, 114, 3885–3892. [CrossRef] [PubMed] [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically eukaryotic 16S-like rRNA-coding regions. Gene, 71, 491–499. [CrossRef] [Google Scholar]
- Mugridge NB, Morrison DA, Heckeroth AR, Johnson AM, Tenter AM. 1999. Phylogenetic analysis based on full-length large subunit ribosomal RNA gene sequence comparison reveals that Neospora caninum is more closely related to Hammondia heydorni than to Toxoplasma gondii. International Journal for Parasitology, 29, 1545–1556. [CrossRef] [PubMed] [Google Scholar]
- Nigro M, Mancianti F, Rossetti P, Poli A. 1991. Ultrastructure of the cyst and life cycle of Sarcocystis sp. from wild sheep (Ovis musimon). Journal of Wildlife Diseases, 27, 217–224. [CrossRef] [PubMed] [Google Scholar]
- Odening K, Stolte M, Bockhardt I. 1996. On the diagnostics of Sarcocystis in chamois (Rupicapra rupicapra). Applied Parasitology, 37, 153–160. [Google Scholar]
- Odening K, Stolte M, Bockhardt I. 1996. Sarcocysts in exotic Caprinae (Bovidae) from zoological gardens. Acta Parasitologica, 41, 67–75. [Google Scholar]
- Odening K, Stolte M, Walter G, Bockhardt I. 1995. Cyst wall ultrastructure of two Sarcocystis spp. from European mouflon (Ovis ammon musimon) in Germany compared with domestic sheep. Journal of Wildlife Diseases, 31, 550–554. [CrossRef] [PubMed] [Google Scholar]
- Saito M, Shibata Y, Kubo M, Itagaki H. 1997. Sarcocystis mihoensis n. sp. from sheep in Japan. Journal of Veterinary Medical Science, 59, 103–106. [CrossRef] [Google Scholar]
- Wang G, Wei T, Wang X, Li W, Zhang P, Dong M, Xiao H. 1988. The morphology and life cycle of Sarcocystis microps n. sp. in sheep of Qinghai in China. China Veterinary Technology, 6, 9–11. [Google Scholar]
Cite this article as: Hu J-J, Huang S, Wen T, Esch GW, Liang Y & Li H-L: Sarcocystis spp. in domestic sheep in Kunming City, China: prevalence, morphology, and molecular characteristics. Parasite, 2017, 24, 30.
All Tables
Prevalence of Sarcocystis spp. in different muscular tissues of sheep (n = 86) in Kunming, China.
All Figures
 |
Figure 1. Morphological characteristics of Sarcocystis tenella sarcocysts from sheep muscle. (A) Light micrograph of a sarcocyst (unstained); note the sarcocyst wall with numerous palisade-like protrusions (pp). Scale bar = 20 μm. (B) Longitudinal section of sarcocyst wall by transmission electron microscopy (TEM). The sarcocyst wall had palisade-like protrusions (pp), characterized by the apex which contained dense plaques (arrow); a layer of ground substances (gs) located beneath the primary sarcocyst wall surrounded bradyzoites (bz). Scale bar = 1 μm. (C) Cross-section of a sarcocyst by TEM; note the dense plaques (arrow), ground substance (gs), bradyzoites (bz), and host cell (hc). Scale bar = 1 μm. |
| In the text | |
 |
Figure 2. Morphological characteristics of Sarcocystis arieticanis sarcocysts from sheep muscle. (A) Light micrograph of a sarcocyst (unstained); note the sarcocyst wall with numerous hair-like protrusions (hp). Scale bar = 20 μm. (B) Transmission electron micrograph of the cyst wall of a sarcocyst; note host cell (hc), bradyzoites (bz), and hair-like protrusions (arrow). Scale bar = 1 μm. |
| In the text | |
 |
Figure 3. Phylogenetic tree for selected members of the Sarcocystidae based on 18S rRNA sequences and inferred by the maximum parsimony (MP) method using the Tree-Bisection-Regrafting (TBR) algorithm. The analysis involved 52 nucleotide sequences (GenBank accession numbers behind the taxon names), and a total of 2,014 positions in the final dataset. The values between the branches represent percent bootstrap value per 1,000 replicates, and the values below 50% are not shown. The three new sequences of Sarcocystis tenella (MF039329) and S. arieticanis (MF039330 and MF039331) have the taxon name in boldface. |
| In the text | |
 |
Figure 4. Phylogenetic tree for selected members of the Sarcocystidae based on 28S rRNA sequences and inferred by the maximum parsimony (MP) method using the Tree-Bisection-Regrafting (TBR) algorithm. The analysis involved 20 nucleotide sequences (GenBank accession numbers behind the taxon names), and a total of 4,517 positions in the final dataset. The values between the branches represent percent bootstrap value per 1,000 replicates, and the values below 50% are not shown. The four new sequences of Sarcocystis tenella (MF039325 and MF039326) and S. arieticanis (MF039327 and MF039328) have the taxon name in boldface. |
| In the text | |
 |
Figure 5. Phylogenetic tree for selected members of the Sarcocystidae based on cox1 sequences and inferred by the maximum parsimony (MP) method using the Tree-Bisection-Regrafting (TBR) algorithm. The analysis involved 27 nucleotide sequences (GenBank accession numbers behind the taxon names), and a total of 998 positions in the final dataset. The values between the branches represent percent bootstrap value per 1,000 replicates, and the values below 50% are not shown. The three new sequences of Sarcocystis tenella (MF039322 and MF039323) and S. arieticanis (MF039324) have the taxon name in boldface. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


