| Issue |
Parasite
Volume 31, 2024
|
|
|---|---|---|
| Article Number | 34 | |
| Number of page(s) | 9 | |
| DOI | https://doi.org/10.1051/parasite/2024033 | |
| Published online | 01 July 2024 | |
Research Article
Cross-species transmission of Cryptosporidium in wild rodents from the southern region of Zhejiang Province of China and its possible impact on public health
Transmission interspécifique de Cryptosporidium chez les rongeurs sauvages de la région sud de la province chinoise du Zhejiang et son impact possible sur la santé publique
1
National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention (Chinese Center for Tropical Diseases Research), National Key Laboratory of Intelligent Tracking and Forecasting for Infectious Diseases, NHC Key Laboratory of Parasite and Vector Biology, National Center for International Research on Tropical Diseases, WHO Collaborating Centre for Tropical Diseases, 200025 Shanghai, China
2
Department of Parasitology, School of Basic Medical Sciences, Wenzhou Medical University, Wenzhou, Zhejiang 325035, China
3
Department of Pathogenic Biology, Hainan Medical University, Haikou, Hainan, China
4
Hainan Medical University – The University of Hong Kong Joint Laboratory of Tropical Infectious Diseases, Hainan Medical University, Haikou, Hainan, China
5
Key Laboratory of Tropical Translational Medicine of Ministry of Education, Hainan Medical University, Haikou 571199, China
* Corresponding authors: jiangyy@nipd.chinacdc.cn (Yanyan Jiang); ganglu2018@163.com (Gang Lu); hhc@wmu.edu.cn (Huicong Huang); hayidazhaowei@163.com (Wei Zhao)
Received:
6
March
2024
Accepted:
31
May
2024
Wild rodents serve as reservoirs for Cryptosporidium and are overpopulated globally. However, genetic data regarding Cryptosporidium in these animals from China are limited. Here, we have determined the prevalence and genetic characteristics of Cryptosporidium among 370 wild rodents captured from three distinct locations in the southern region of Zhejiang Province, China. Fresh feces were collected from the rectum of each rodent, and DNA was extracted from them. The rodent species was identified by PCR amplifying the vertebrate cytochrome b gene. Cryptosporidium was detected by PCR amplification and amplicon sequencing the small subunit of ribosomal RNA gene. Positive samples of C. viatorum and C. parvum were further subtyped by analyzing the 60-kDa glycoprotein gene. A positive Cryptosporidium result was found in 7% (26/370) of samples, involving five rodent species: Apodemus agrarius (36), Niviventer niviventer (75), Rattus losea (18), R. norvegicus (155), and R. tanezumi (86). Their respective Cryptosporidium positive rates were 8.3%, 5.3%, 11.1%, 7.1%, and 7.0%. Sequence analysis confirmed the presence of three Cryptosporidium species: C. parvum (4), C. viatorum (1), and C. muris (1), and two genotypes: Cryptosporidium rat genotype IV (16) and C. mortiferum-like (4). Additionally, two subtypes of C. parvum (IIdA15G1 and IIpA19) and one subtype of C. viatorum (XVdA3) were detected. These results demonstrate that various wild rodent species in Zhejiang were concurrently infected with rodent-adapted and zoonotic species/genotypes of Cryptosporidium, indicating that these rodents can play a role in maintaining and dispersing this parasite into the environment and other hosts, including humans.
Résumé
Les rongeurs sauvages servent de réservoirs à Cryptosporidium et ont des grandes populations à l’échelle mondiale. Cependant, les données génétiques concernant Cryptosporidium chez ces animaux en Chine sont limitées. Ici, nous avons déterminé la prévalence et les caractéristiques génétiques de Cryptosporidium parmi 370 rongeurs sauvages capturés dans trois endroits distincts de la région sud de la province du Zhejiang, en Chine. Des excréments frais ont été collectés dans le rectum de chaque rongeur et l’ADN en a été extrait. L’espèce de rongeur a été identifiée par amplification par PCR du gène du cytochrome b des vertébrés. Cryptosporidium a été détecté par amplification PCR et séquençage d’amplicons de la petite sous-unité du gène de l’ARN ribosomal. Les échantillons positifs de C. viatorum et C. parvum ont ensuite été sous-typés en analysant le gène de la glycoprotéine de 60 kDa. Un résultat positif pour Cryptosporidium a été trouvé dans 7 % (26/370) des échantillons, impliquant cinq espèces de rongeurs : Apodemus agrarius (36), Niviventer niviventer (75), Rattus losea (18), R. norvegicus (155) et R. tanezumi (86). Leurs taux respectifs de positivité pour Cryptosporidium étaient de 8,3 %, 5,3 %, 11,1 %, 7,1 % et 7,0 %. L’analyse des séquences a confirmé la présence de trois espèces de Cryptosporidium : C. parvum (4), C. viatorum (1) et C. muris (1), et de deux génotypes : Cryptosporidium génotype IV de rat (16) et C. mortiferum-like (4). De plus, deux sous-types de C. parvum (IIdA15G1 et IIpA19) et un sous-type de C. viatorum (XVdA3) ont été détectés. Ces résultats démontrent que diverses espèces de rongeurs sauvages du Zhejiang sont simultanément infectées par des espèces/génotypes de Cryptosporidium zoonotiques et adaptés aux rongeurs, ce qui indique que ces rongeurs peuvent jouer un rôle dans le maintien et la dispersion de ce parasite dans l’environnement et d’autres hôtes, y compris les humains.
Key words: Cryptosporidium / Molecular detection / Wild rodents / Zoonotic / Public health, China
© Y. Jiang et al., published by EDP Sciences, 2024
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Cryptosporidium, a protozoan parasite that colonizes the intestines, is a significant contributor to moderate to chronic diarrhea and related fatalities among children under two years of age and immunocompromized patients (HIV-positive) [4, 34]. Additionally, Cryptosporidium has been identified as able to infect over 260 species of animals [24]. Humans can acquire this parasite through various routes, encompassing direct contact with infected individuals or animals, and ingestion of contaminated water and food [35]. The public health importance of cryptosporidiosis became evident with the global recognition of Cryptosporidium as the predominant waterborne parasite [8]. To effectively minimize the frequency of Cryptosporidium outbreaks, it is imperative to identify potential sources of infection and likely modes of transmission [13]. Thus, monitoring Cryptosporidium in different hosts becomes critical, especially in animal hosts in close contact with humans.
A wide range of molecular epidemiological strategies have been used to characterize Cryptosporidium species at species/genotype and subtype levels [24]. Currently, this parasite has been identified with an estimated 120 genotypes and 50 valid species [23, 26]. Moreover, almost 21 distinct species/genotypes of this parasite have been found in humans, primarily as a result of zoonotic transmission, where the infection is transmitted from animals to humans [23]. Rodents, as a key reservoir of Cryptosporidium, have attracted widespread attention, particularly wild ones, considering their involvement alongside various animals (domestic, stray, and wild) and water sources in maintaining the stability and continuity of the Cryptosporidium transmission cycle. The presented data indicate that rodents harbor a minimum of 25 species and 48 genotypes of Cryptosporidium. Among these, wild rodents harbor 21 species and 32 genotypes, with C. parvum being the most common species [36]. Therefore, wild rodents potentially play a pivotal role in the transmission of zoonotic Cryptosporidium species. Despite this understanding, significant gaps exist concerning the incidence of Cryptosporidium infection in various nations and territories. For instance, in China, molecular studies on Cryptosporidium species in wild rodent species have been restricted to a small number of species [15, 22, 36].
Cryptosporidium species have been observed to exhibit a high prevalence in diverse animal species, including pigs, cattle, chickens, and horses, within the geographical region of Zhejiang Province, China [9, 28, 33, 40]. Moreover, they have also been found in patients with diarrhea, as well as in the source water of several cities of this province [2, 21, 37]. However, currently, there is only one study that confirms the presence of this parasite in R. norvegicus from Jiaxing City in Zhejiang [22]. The objective of the present study was to investigate the distribution, prevalence, and genetic characteristics of Cryptosporidium species among wild rodents residing in southern Zhejiang Province.
Materials and methods
Ethics
The present study was conducted as per the recommendations of the Chinese Laboratory Animal Administration Act (1988), which regulates the ethical handling and use of animals in scientific studies. All protocols were carefully examined and approved by the Research Ethics Committee of Wenzhou Medical University (SCILLSC-2021-01).
Sample collection
A total of 370 wild rodents were trapped in three distinct locations within rural areas immediately adjacent to human habitations in Zhejiang Province, between April 1 and October 14, 2023 (specifically, encompassing the second week of April, June, August, and October in the year 2023) (Fig. 1). Among these, 68 were caught in Yueqing (Hongqiao), 102 in Yongjia (Yantou), and 200 in Rui’an (Tangxia, Pandai, Shangwang). All wild rodents were trapped in cage traps baited with deep-fried dough sticks. In each designated location, around 50 cage traps were deployed at sunset and collected before sunrise. The traps were positioned in a linear setup, with 5 m between each trap, forming transects. All rodents were shifted to the controlled laboratory environment within 48 h following their capture and euthanized via CO2 inhalation. Data related to the collection time and region was noted after these rodents were captured via trapping. Following that, a fresh feces sample (500 mg) was obtained immediately from the rectum of each rodent. The sample was then stored in ice boxes and shifted to the laboratory, where its DNA was extracted within a week.
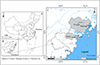 |
Figure 1 Map of rodent sampling locations in Wenzhou, Zhejiang Province, China. The figure was originally designed by the authors under ArcGIS 10.4 software. The original vector diagram imported in ArcGIS was adapted from the National Geomatics Center of China (http://www.ngcc.cn). The map has been modified and assembled according to permission and attribution guidelines. |
DNA extraction
As per the manufacturer’s recommendations, genomic DNA was isolated from each processed sample (200 mg) via a QIAamp DNA Mini Stool Kit (QIAGEN, Hilden, Germany). To achieve a significant yield of DNA, the lysate temperature was elevated to 95 °C. Before the PCR analysis, the DNA reconstituted in 200 μL of AE elution buffer (supplied with the kit) was kept at −20 °C.
Identification of rodent species
The rodent species were identified via PCR amplification of the vertebrate cytochrome b (cytb) gene with 421 bp amplified from fecal DNA. The primer design and PCR conditions were in line with the guidelines defined by Verma and Singh (2003) [27]. Each PCR reaction was comprised of 35 cycles, which included denaturation at 94 °C for 30 s, annealing at 51 °C for 30 s, and extension at 72 °C for 30 s. An initial denaturation step was also performed at 94 °C for 5 min, followed by the completion of a final extension at 72 °C for 5 min.
Cryptosporidium genotyping and subtyping
Nested PCR was performed on all isolated DNA with a specific target, using an 830 bp fragment of the partial small subunit of ribosomal RNA (SSU rRNA) gene of Cryptosporidium for amplification. Based on a previous description, primers were synthesized [32]. The 60-kDa glycoprotein (gp60) gene was amplified using nested PCR, enabling the further subtyping of positive isolates of C. parvum and C. viatorum using the same primers previously designed by Alves et al. (2003) and Stensvold et al. (2015), respectively [1, 25]. In every PCR amplification process, TaKaRa Taq DNA Polymerase (TaKaRa Bio Inc., Tokyo, Japan) was utilized. To ensure the validity of the reactions, positive controls, which contain C. bailey DNA derived from chickens, and negative controls, where no DNA template is included, were incorporated in every PCR reaction. Before sequencing, secondary PCR products were observed on 1.5% agarose gels, followed by staining with GelRed (Biotium, Fremont, CA, USA).
Sequencing and phylogenetic analysis
The commercial sequencing of amplified products of SSU rRNA and gp60 genes of Cryptosporidium spp. was performed by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). Two-way sequencing was used to validate the accuracy of the sequence. To examine the species and subtype of Cryptosporidium species, the identified sequences were aligned with the reference sequences obtained from the National Center for Biotechnology (https://www.ncbi.nlm.nih.gov/) using ClustalX 2.0 (http://www.clustal.org/). In MEGA 11, a neighbor-joining (NJ) method with a Kimura 2-parameter model was used to conduct phylogenetic analyses, with the objective of assessing the phylogenetic relationships among the sequences obtained in this study and pertinent reference sequences available in GenBank. The clusters’ stability was evaluated using 1000 replicates and Bootstrap analysis.
Statistical analyses
Data analysis was performed with SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). The chi-square test was utilized to compare the prevalence of Cryptosporidium spp. between areas, gender, rodent species and season groups, respectively. A p-value < 0.05 was considered indicative of statistical significance.
Nucleotide sequence accession numbers
The nucleotide sequences of Cryptosporidium obtained in this study were deposited in the GenBank database under accession numbers PP038021 to PP038023 and PP038026 to PP038028 for SSU rRNA, and PP104938 to PP104940 for gp60.
Results
Study population
This study used PCR and sequencing analysis of the cytb gene to identify five species of rodents, including Rattus norvegicus (n = 155), R. tanezumi (n = 86), Niviventer niviventer (n = 75), Apodemus agrarius (n = 36) and R. losea (n = 18) (Table 1 and Table S1). The majority of samples were obtained in the summer (43.2%, 160/370), then in spring (32.2%, 119/370) and autumn (24.6%, 91/370); none were collected in the winter. The sex of the rodents was reported as 52.7% (195/370) females and 47.3% (175/370) males (Table 1 and Table S1).
Prevalence and species/genotype of Cryptosporidium in the investigated rodent by species, season, gender, and location.
Prevalence of Cryptosporidium infection
Nested PCR was conducted on 370 fecal samples to evaluate the existence of Cryptosporidium species via the SSU rRNA gene. A total of 26 samples tested positive for this parasite with an average (7.0%) infection rate. Cryptosporidium existed in all three areas, with infection rates of 14.7% (Yeqing), 2.9% (Yongjia), and 6.5% (Rui’an) (Table 1 and Table S1). Statistical analysis revealed significant variations in the prevalence of Cryptosporidium among the three regions (χ2 = 8.829; df = 2; p = 0.012).
The infection rates of Cryptosporidium vary among rodent species, ranging from 5.3% (4/75) in N. niviventer to 11.1% (2/18) in R. losea, with 7.1% (11/155) in R. norvegicus, 7.0% (6/86) in R. tanezumi, and 8.3% (3/36) in A. agrarius (Table 1 and Table S1). The highest detection rate of Cryptosporidium in rodents collected in spring, reaching 9.2% (11/119), followed by 7.7% (7/91) in autumn, and 5.5% (8/160) in summer (Table 1 and Table S1). However, the difference between infection rates of Cryptosporidium in the groups of rodent species and seasons was not regarded as statistically significant (p > 0.05). In relation to gender, the incidence of Cryptosporidium was comparatively lower in female (4.6%) (9/195) than in male (9.7%; 17/175) rodents, but without statistical significance (χ2 = 3.67; df = 1; p = 0.06) (Table 1 and Table S1).
Cryptosporidium species/genotypes distribution
Five Cryptosporidium species or genotypes were detected, including Cryptosporidium rat genotype IV (n = 16), C. parvum (n = 4), C. mortiferum-like genotype (n = 4), C. viatorum (n = 1), and C. muris (n = 1) (Table 1 and Table S1). The prevalence of Cryptosporidium rat genotype IV was observed to be predominant (61.5%; 16/26) among the wild rodent population. This genotype was detected in four out of the five rodent species, except A. agrarius (Table 1 and Table S1). Of the four C. parvum isolates, two each were found in R. norvegicus and A. agrarius. Cryptosporidium mortiferum-like was only found in R. norvegicus, while C. viatorum and C. muris were identified in a single R. norvegicus and A. agrarius, respectively (Table 1 and Table S1).
The sampling sites exhibit differences in the distribution of Cryptosporidium species. Specifically, Yeqing yielded the Cryptosporidium rat genotype IV, C. viatorum and C. parvum. Yongjia yielded C. mortiferum-like and Cryptosporidium rat genotype IV. While Cryptosporidium rat genotype IV, C. parvum, C. muris and C. mortiferum-like were discovered in Rui’an (Table 1 and Table S1).
Meanwhile, Cryptosporidium rat genotype IV was detected throughout all three seasons, whereas C. muris was exclusively detected in the summer. Conversely, C. viatorum and C. mortiferum-like were only detected in the spring (Table 1). In terms of gender, both male and female rodents were found to harbor Cryptosporidium rat genotype IV and C. parvum, whereas only female rodents were found to possess C. muris and C. mortiferum-like, and male rodents were found to harbor C. viatorum (Table 1 and Table S1).
Genetic identification of Cryptosporidium species/genotypes
At the SSU rRNA locus, among the 16 sequences of Cryptosporidium rat genotype IV, 10 and six sequences were identical and had 100% similarity with the sequence AY737582 of genotype W19 variant in storm water from the USA and MG917671 of genotype W19 variant in brown rats from China, respectively (Table S2). Four sequences of C. parvum were identical and had 100% similarity with the sequence OM146539 of C. parvum in humans from Sweden, as well as eight other sequences in Macaca mulatta or bamboo rats from China (Table S2). The four C. mortiferum-like isolates possessed identical sequences which have not been documented previously and exhibit a sequence similarity of 98.78% to the C. mortiferum sequence (OP935211) detected in humans from the USA (Table S2). The sequence of C. viatorum was similar and it has not been documented previously and exhibited homology of 99.61% to the sequence MK522270 of C. viatorum isolated in Berylmys bowersi from China (Table S2). The sequence of C. muris exhibited 100% identity to KF419208, which is found in R. norvegicus from China (Table S2). In the phylogenetic tree, the sequences belonging to the same species were shown to form distinct clusters, as depicted in Figure 2.
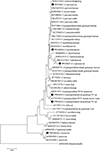 |
Figure 2 Phylogenetic tree of Cryptosporidium species based on SSU rRNA sequences. The tree was generated using a neighbor-joining analysis, with genetic distances calculated via the Kimura 2-parameter model. Bootstrap values (>50%) derived from 1000 replicates are displayed to the left of the nodes for reliability assessment. The sequences generated in the present study are indicated with the solid circles. |
At the gp60 gene locus, successful amplification was achieved for C. viatorum and three C. parvum-positive isolates. Two of the three gp60 sequences obtained from C. parvum exhibited 100% resemblance to the sequence of M. mulatta in China, from which C. parvum subtype IIdA15G1 (KJ917586) was identified (Table S2). The other one, gp60 sequence of C. parvum, shared 100% similarity to subtype IIpA6 (MK956001) from bamboo rats in China (Table S2). The C. viatorum sample had the same gp60 sequence which has not been documented in the previous literature and possesses a nucleotide similarity of 99.51% with the well-documented subtype XVdA3 (MK433560) of C. viatorum originating from Leopoldamys edwardsi in China (Table S2). Figure 3 presents the phylogenetic tree, showing the genetic correlations among the gp60 subtypes of C. parvum and C. viatorum.
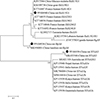 |
Figure 3 Phylogenetic relationships of gp60 subtypes of C. parvum and C. viatorum identified in the study and other gp60 subtype sequences deposited in GenBank, as inferred by a neighbor-joining analysis of gp60 sequences based on the genetic distance by the Kimura 2-parameter model. The numbers displayed on the branches represent the percentage bootstrapping outcomes derived from 1000 replicates. The sequences generated in the present study are indicated with solid circles. |
Discussion
In the present study, the average rate of Cryptosporidium infection among the identified rodents was 7.0%, which was found to be lower than the aggregated global rate for wild rodents (20.5%), as determined by Zhang et al. [36]. In China, Cryptosporidium has been found in a variety of rodents, where the infection rates vary by their types, such as 4.0–73.9% in wild rodents, 2.1–29.5% in farmed rodents, 0.6–8.6% in lab rodents and 1.4–100% in pet rodents [22, 36]. The differences in rodent species, detection strategies, animal age, sample size, and study locations might be responsible for the disparity in prevalence.
The present study identified five Cryptosporidium species/genotypes including Cryptosporidium rat genotype IV, C. mortiferum-like, C. parvum, C. viatorum and C. muris. Multiple studies have demonstrated that rats serve as a predominant host species for Cryptosporidium rat genotype IV (formerly known as Cryptosporidium environmental sequence, Cryptosporidium genotypes W19, or W19 variant) [36, 39]. Cryptosporidium rat genotype IV has previously been found in Asian house rats, Edward’s long-tailed rats, Muridae, and brown rats from China [6, 38], and it has also been identified in rats from Japan, Spain and Sweden [3, 14, 17]. However, despite the discovery of Cryptosporidium rat genotype IV in Asiatic black bears and cats from China and in one-humped camels from Egypt, limited data exist regarding the chances for infection of humans and other animals by Cryptosporidium rat genotype IV [7, 19, 29]. Consequently, the possibility of this genotype inducing disease in livestock or humans remains uncertain. Further systemic molecular epidemiological studies into Cryptosporidium species with a wider range of hosts are required in the future to identify the exact host distribution of Cryptosporidium rat genotype IV.
Zoonotic species include C. muris, C. viatorum and C. parvum, due to the extensive documentation of their infections in humans and a diverse array of mammalian hosts [23]. For example, C. parvum, which is prevalent in rodents worldwide, has been consistently identified in wildlife, having infected over 40 species of wild animals [23, 36]. In China 16.7% of human cases (44/263) of cryptosporidiosis were attributed to C. parvum, a prevalent pathogen in farmed animals, including cattle, sheep, and goats [12, 20]. Further, 18.7% of rodent-derived Cryptosporidium cases (189/1010) had been confirmed to be caused by C. parvum [36]. Initially, C. viatorum was detected in travelers from the Indian subcontinent who had arrived in the United Kingdom [7]. More than 13 countries, including China, have reported cases of C. viatorum in humans [25, 31]. Further analysis revealed the presence of C. viatorum in several rodent species, including R. rattus from France, R. lutreolus from Australia and Leopoldamys edwardsi and Berylmys bowersi from China [10, 16, 38]. Additionally, C. muris has been extensively documented in various mammalian hosts, such as rodents, felids, canids, equids, suids, non-human primates, etc. [23]. The transmission of C. parvum, C. viatorum and C. muris from wild rodent species to humans and other animals via cross-species contact could, therefore, not be ignored.
This study identified a novel genotype in R. norvegicus that shares genetic similarities to C. mortiferum (Cryptosporidium chipmunk genotype I), named C. mortiferum-like. The sequences of C. mortiferum-like discovered in this study have not been previously reported in the literature. However, it is well known that C. mortiferum can infect people, and several human cases have been reported [11, 23, 26]. Therefore, C. mortiferum-like is highly likely to also have the ability to infect people, and of course, clear evidence needs to be provided through more research in the future. The discovery of novel sequences of Cryptosporidium in R. norvegicus suggests the existence of some novel Cryptosporidium species/genotypes in wild rodents. This is primarily due to the order of rodents having the most diverse of all mammalian groups.
In the evaluation of C. parvum in both animals and humans, subtyping tools are frequently used. The transmission of C. parvum between animals and humans was enhanced via the application of subtype-specific molecular diagnostic tools [12, 24]. At least 15 subtype families for C. parvum were detected via gp60 gene analysis, including IIa-IIi and IIk-IIp [12]. In China, at least 20 subtypes have been identified, with IIaA15G2R1, IIaA15G2R2, IIaA13G2R2, IIdA15G1 and IIdA14G1 being found in humans [12, 20]. Wild rodents were examined in the present study that observed two subtypes (IIdA15G1 and IIpA6) of C. parvum. The subtype IIdA15G1 is one of the prevalent subtypes found in cattle, exhibiting a diverse geographic distribution across China [12]. Its elevated mortality rate among pre-weaned dairy calves in China has been attributed to multiple outbreaks of cryptosporidiosis [5, 12]. Furthermore, the subtype was subsequently detected in non-human primates and humans in China, which provides further evidence for the possibility of zoonotic transmission [12, 30, 37]. Thus, IIdA15G1-infected wild rodents pose a potential risk to both humans and other animals. However, IIpA6 has only been detected in bamboo rodents thus far, and its potential to infect humans and livestock is unknown [18]. A comprehensive understanding of the host range of subtype IIpA6 of Cryptosporidium species would require systematic molecular epidemiological studies across a wider range of hosts.
Conclusions
The present study provided evidence of the presence of Cryptosporidium in five species of wild rodents in Zhejiang, China, with an average infection rate of 7.0%. The presented molecular findings suggest that Cryptosporidium rat genotype IV predominantly infect wild rodents. As a result, these rodents have a restricted capacity to serve as natural reservoirs for human infections. In contrast, the discovery of C. muris, C. parvum, C. mortiferum-like, C. viatorum and C. viatorum suggests a connection between rodents and humans. This finding demonstrates that animals infected with Cryptosporidium have substantial zoonotic potential and indicates that wild rodents could serve as a reservoir for human cryptosporidiosis caused by the Cryptosporidium species above.
Funding
This work was supported by the National Key Research and Development Program of China (grant number 2021YFC2300902 to YJ), the National Natural Science Foundation of China (82273693 to YJ) and the Basic scientific research project of Wenzhou (Y2023070 to WZ). The funding sponsors had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors do not have a commercial or other association that represents a conflict of interest.
Supplementary Tables
Table S1: Prevalence and species of Cryptosporidium in the investigated wild rodents by species, season, gender, and location.
Table S2: Similarity analysis of SSUrRNA and Gp60 sequences of Cryptosporidium obtained in this study.
Access hereReferences
- Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. Journal of Clinical Microbiology, 41(6), 2744–2747. [CrossRef] [PubMed] [Google Scholar]
- An W, Zhang D, Xiao S, Yu J, Yang M. 2011. Quantitative health risk assessment of Cryptosporidium in rivers of southern China based on continuous monitoring. Environmental Science & Technology, 45(11), 4951–4958. [CrossRef] [PubMed] [Google Scholar]
- Backhans A, Jacobson M, Hansson I, Lebbad M, Lambertz ST, Gammelgård E, Saager M, Akande O, Fellström C. 2013. Occurrence of pathogens in wild rodents caught on Swedish pig and chicken farms. Epidemiology and Infection, 141(9), 1885–1891. [CrossRef] [PubMed] [Google Scholar]
- Bouzid M, Hunter PR, Chalmers RM, Tyler KM. 2013. Cryptosporidium pathogenicity and virulence. Clinical Microbiology Reviews, 26(1), 115–134. [CrossRef] [PubMed] [Google Scholar]
- Cui Z, Wang R, Huang J, Wang H, Zhao J, Luo N, Li J, Zhang Z, Zhang L. 2014. Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasites & Vectors, 7, 529. [CrossRef] [PubMed] [Google Scholar]
- Deng L, Chai Y, Luo R, Yang L, Yao J, Zhong Z, Wang W, Xiang L, Fu H, Liu H, Zhou Z, Yue C, Chen W, Peng G. 2020. Occurrence and genetic characteristics of Cryptosporidium spp. and Enterocytozoon bieneusi in pet red squirrels (Sciurus vulgaris) in China. Scientific Reports, 10(1), 1026. [CrossRef] [PubMed] [Google Scholar]
- Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM. 2012. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. International Journal for Parasitology, 42(7), 675–682. [CrossRef] [PubMed] [Google Scholar]
- Fayer R. 2004. Cryptosporidium: A water-borne zoonotic parasite. Veterinary Parasitology, 126(1–2), 37–56. [CrossRef] [PubMed] [Google Scholar]
- Feng X, Deng J, Zhang Z, Yu F, Zhang J, Shi T, Sun H, Qi M, Liu X. 2023. Dominant infection of Cryptosporidium baileyi in broiler chickens in Zhejiang Province, China. Parasitology Research, 122(9), 1993–2000. [CrossRef] [PubMed] [Google Scholar]
- García-Livia K, Fernández-Álvarez Á, Feliu C, Miquel J, Quilichini Y, Foronda P. 2022. Cryptosporidium spp. in wild murids (Rodentia) from Corsica, France. Parasitology Research, 121(1), 345–354. [CrossRef] [PubMed] [Google Scholar]
- Guo Y, Cebelinski E, Matusevich C, Alderisio KA, Lebbad M, McEvoy J, Roellig DM, Yang C, Feng Y, Xiao L. 2015. Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. Journal of Clinical Microbiology, 53(5), 1648–1654. [CrossRef] [PubMed] [Google Scholar]
- Guo Y, Ryan U, Feng Y, Xiao L. 2022. Emergence of zoonotic Cryptosporidium parvum in China. Trends in Parasitology, 38(4), 335–343. [CrossRef] [PubMed] [Google Scholar]
- Helmy YA, Hafez HM. 2022. Cryptosporidiosis: From prevention to treatment, a narrative review. Microorganisms, 10(12), 2456. [CrossRef] [PubMed] [Google Scholar]
- Hikosaka K, Nakai Y. 2005. A novel genotype of Cryptosporidium muris from large Japanese field mice, Apodemus speciosus. Parasitology Research, 97(5), 373–379. [CrossRef] [PubMed] [Google Scholar]
- Hu B, Wang J, Zhang S, Wang B, Xing Y, Han S, He H. 2022. Novel genotypes of Cryptosporidium and Enterocytozoon bieneusi detected in plateau zokors (Myospalax baileyi) from the Tibetan Plateau. International Journal for Parasitology: Parasites and Wildlife, 19, 263–268. [CrossRef] [Google Scholar]
- Koehler AV, Wang T, Haydon SR, Gasser RB. 2018. Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus – An emerging zoonotic pathogen?. International Journal for Parasitology: Parasites and Wildlife, 7(1), 18–26. [CrossRef] [Google Scholar]
- Köster PC, Dashti A, Bailo B, Muadica AS, Maloney JG, Santín M, Chicharro C, Migueláñez S, Nieto FJ, Cano-Terriza D, García-Bocanegra I, Guerra R, Ponce-Gordo F, Calero-Bernal R, González-Barrio D, Carmena D. 2021. Occurrence and genetic diversity of protist parasites in captive non-human primates, zookeepers, and free-living sympatric rats in the Córdoba zoo conservation centre, Southern Spain. Animals, 11(3), 700. [CrossRef] [PubMed] [Google Scholar]
- Li F, Zhao W, Zhang C, Guo Y, Li N, Xiao L, Feng Y. 2020. Cryptosporidium species and C. parvum subtypes in farmed bamboo rats. Pathogens, 9(12), 1018. [CrossRef] [PubMed] [Google Scholar]
- Li J, Dan X, Zhu K, Li N, Guo Y, Zheng Z, Feng Y, Xiao L. 2019. Genetic characterization of Cryptosporidium spp. and Giardia duodenalis in dogs and cats in Guangdong, China. Parasites & Vectors, 12(1), 571. [CrossRef] [PubMed] [Google Scholar]
- Liu A, Gong B, Liu X, Shen Y, Wu Y, Zhang W, Cao J. 2020. A retrospective epidemiological analysis of human Cryptosporidium infection in China during the past three decades (1987–2018). PLoS Neglected Tropical Diseases, 14(3), e0008146. [CrossRef] [PubMed] [Google Scholar]
- Liu H, Ni H, Liu S, Shen Y, Wang R, Cao J, Yin J. 2023. First report on occurrence and genotypes of Enterocytozoon bieneusi, Cryptosporidium spp. and Cyclospora cayetanensis from diarrheal outpatients in Ningbo, Southeast China. Microbial Pathogenesis, 174, 105952. [CrossRef] [PubMed] [Google Scholar]
- Ni HB, Sun YZ, Qin SY, Wang YC, Zhao Q, Sun ZY, Zhang M, Yang D, Feng ZH, Guan ZH, Qiu HY, Wang HX, Xue NY, Sun HT. 2021. Molecular detection of Cryptosporidium spp. and Enterocytozoon bieneusi infection in wild rodents from six provinces in China. Frontiers in Cellular and Infection Microbiology, 11, 783508. [CrossRef] [PubMed] [Google Scholar]
- Ryan U, Zahedi A, Feng Y, Xiao L. 2021. An update on zoonotic Cryptosporidium species and genotypes in humans. Animals, 11(11), 3307. [CrossRef] [PubMed] [Google Scholar]
- Ryan UM, Feng Y, Fayer R, Xiao L. 2021. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia – A 50 year perspective (1971–2021). International Journal for Parasitology, 51(13–14), 1099–1119. [CrossRef] [PubMed] [Google Scholar]
- Stensvold CR, Elwin K, Winiecka-Krusnell J, Chalmers RM, Xiao L, Lebbad M. 2015. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. Journal of Clinical Microbiology, 53(6), 1891–1897. [CrossRef] [PubMed] [Google Scholar]
- Tůmová L, Ježková J, Prediger J, Holubová N, Sak B, Konečný R, Květoňová D, Hlásková L, Rost M, McEvoy J, Xiao L, Santín M, Kváč M. 2023. Cryptosporidium mortiferum n. sp. (Apicomplexa: Cryptosporidiidae), the species causing lethal cryptosporidiosis in Eurasian red squirrels (Sciurus vulgaris). Parasites & Vectors, 16(1), 235. [CrossRef] [PubMed] [Google Scholar]
- Verma SK, Singh L. 2003. Novel universal primers establish identity of an enormous number of animal species for forensic application. Molecular Ecology Notes, 3, 28–31. [CrossRef] [Google Scholar]
- Wang L, Xue X, Li J, Zhou Q, Yu Y, Du A. 2014. Cryptosporidiosis in broiler chickens in Zhejiang Province, China: Molecular characterization of oocysts detected in fecal samples. Parasite, 21, 36. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Wang SN, Sun Y, Zhou HH, Lu G, Qi M, Liu WS, Zhao W. 2020. Prevalence and genotypic identification of Cryptosporidium spp. and Enterocytozoon bieneusi in captive Asiatic black bears (Ursus thibetanus) in Heilongjiang and Fujian provinces of China. BMC Veterinary Research, 16(1), 84. [CrossRef] [PubMed] [Google Scholar]
- Wang T, Wei Z, Zhang Y, Zhang Q, Zhang L, Yu F, Qi M, Zhao W. 2022. Molecular detection and genetic characterization of Cryptosporidium in kindergarten children in Southern Xinjiang, China. Infection Genetics and Evolution, 103, 105339. [CrossRef] [Google Scholar]
- Wu Y, Gong B, Liu X, Jiang Y, Cao J, Yao L, Li H, Liu A, Shen Y. 2020. Identification of uncommon Cryptosporidium viatorum (a novel subtype XVcA2G1c) and Cryptosporidium andersoni as well as common Giardia duodenalis assemblages A and B in Humans in Myanmar. Frontiers in Cellular and Infection Microbiology, 10, 614053. [CrossRef] [PubMed] [Google Scholar]
- Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, Fayer R, Lal AA. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Applied and Environmental Microbiology, 65(4), 1578–1583. [CrossRef] [PubMed] [Google Scholar]
- Xu C, Wei Z, Tan F, Liu A, Yu F, Zhao A, Zhang L, Qi M, Zhao W. 2023. Molecular detection and genetic characteristics of Cryptosporidium spp. in Chinese racehorses. Equine Veterinary Journal, 55(3), 474–480. [CrossRef] [PubMed] [Google Scholar]
- Yang X, Guo Y, Xiao L, Feng Y. 2021. Molecular epidemiology of human cryptosporidiosis in low- and middle-income countries. Clinical Microbiology Reviews, 34(2), e00087-19. [CrossRef] [PubMed] [Google Scholar]
- Zahedi A, Ryan U. 2020. Cryptosporidium – An update with an emphasis on foodborne and waterborne transmission. Research in Veterinary Science, 132, 500–512. [CrossRef] [PubMed] [Google Scholar]
- Zhang K, Fu Y, Li J, Zhang L. 2021. Public health and ecological significance of rodents in Cryptosporidium infections. One Health, 14, 100364. [Google Scholar]
- Zhao W, Ren G, Jiang W, Wang L, Wang J, Yuan Z, Yan L, Li Y, Sun Y, Xue X, Jiang Y, Lu G, Huang H. 2024. Genetic characterizations of Cryptosporidium spp. from children with or without diarrhea in Wenzhou, China: High probability of zoonotic transmission. BMC Microbiology, 24(1), 113. [CrossRef] [PubMed] [Google Scholar]
- Zhao W, Zhou H, Huang Y, Xu L, Rao L, Wang S, Wang W, Yi Y, Zhou X, Wu Y, Ma T, Wang G, Hu X, Peng R, Yin F, Lu G. 2019. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: Molecular detection, species/genotype identification and implications for public health. International Journal for Parasitology: Parasites and Wildlife, 9, 317–321. [CrossRef] [Google Scholar]
- Zhao W, Wang J, Ren G, Yang Z, Yang F, Zhang W, Xu Y, Liu A, Ling H. 2018. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasites & Vectors, 11(1), 313. [CrossRef] [PubMed] [Google Scholar]
- Zou Y, Ma JG, Yue DM, Zheng WB, Zhang XX, Zhao Q, Zhu XQ. 2017. Prevalence and risk factors of Cryptosporidium infection in farmed pigs in Zhejiang, Guangdong, and Yunnan provinces, China. Tropical Animal Health and Production, 49(3), 653–657. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Jiang Y, Jiang A, Ren G, Wang L, Xin X, Yuan Z, Liu J, Li Z, Sun Y, Zhou S, Lu G, Huang H & Zhao W. 2024. Cross-species transmission of Cryptosporidium in wild rodents from the southern region of Zhejiang Province of China and its possible impact on public health. Parasite 31, 34.
All Tables
Prevalence and species/genotype of Cryptosporidium in the investigated rodent by species, season, gender, and location.
All Figures
 |
Figure 1 Map of rodent sampling locations in Wenzhou, Zhejiang Province, China. The figure was originally designed by the authors under ArcGIS 10.4 software. The original vector diagram imported in ArcGIS was adapted from the National Geomatics Center of China (http://www.ngcc.cn). The map has been modified and assembled according to permission and attribution guidelines. |
| In the text | |
 |
Figure 2 Phylogenetic tree of Cryptosporidium species based on SSU rRNA sequences. The tree was generated using a neighbor-joining analysis, with genetic distances calculated via the Kimura 2-parameter model. Bootstrap values (>50%) derived from 1000 replicates are displayed to the left of the nodes for reliability assessment. The sequences generated in the present study are indicated with the solid circles. |
| In the text | |
 |
Figure 3 Phylogenetic relationships of gp60 subtypes of C. parvum and C. viatorum identified in the study and other gp60 subtype sequences deposited in GenBank, as inferred by a neighbor-joining analysis of gp60 sequences based on the genetic distance by the Kimura 2-parameter model. The numbers displayed on the branches represent the percentage bootstrapping outcomes derived from 1000 replicates. The sequences generated in the present study are indicated with solid circles. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


