Figure S1:
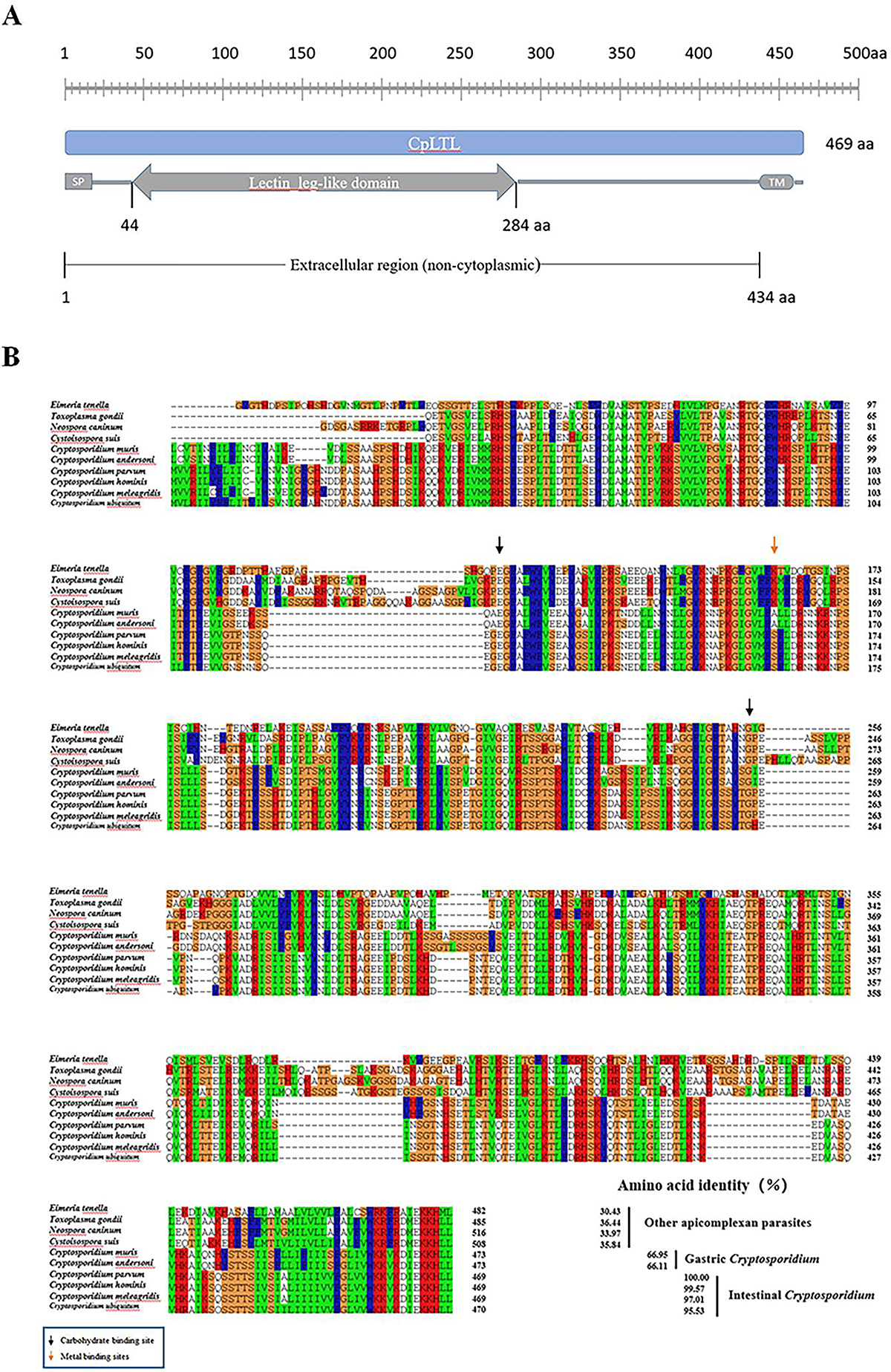
Download original image
Sequence analyses of CpLTL. (A) Domain organization of CpLTL protein using SMART. SP: signal peptide, TM: transmembrane domain. (B) Amino acid sequences of apicomplexan parasite LTLs were aligned with CpLTL using the Clustal W algorithm. The metal-binding sites were marked with a red arrow, and the carbohydrate-binding sites were marked with a black arrow. Amino acid identities with orthologs from other Cryptosporidium species and apicomplexan parasites.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


