| Issue |
Parasite
Volume 29, 2022
|
|
|---|---|---|
| Article Number | 15 | |
| Number of page(s) | 7 | |
| DOI | https://doi.org/10.1051/parasite/2022018 | |
| Published online | 22 March 2022 | |
Research Article
First detection and genotyping of Enterocytozoon bieneusi in pet golden hamsters (Mesocricetus auratus) and Siberian hamsters (Phodopus sungorus) in China
Première détection et génotypage d’Enterocytozoon bieneusi chez des hamsters dorés de compagnie (Mesocricetus auratus) et des hamsters sibériens (Phodopus sungorus) en Chine
College of Animal Science and Technology, Henan University of Science and Technology, No. 263 Kaiyuan Road, Luoyang 471003, China
* Corresponding author: qwf2012@yeah.net
Received:
8
February
2022
Accepted:
8
March
2022
Enterocytozoon bieneusi, a common opportunistic pathogen, has been detected in humans and a wide range of animals worldwide. However, no information on the prevalence and molecular characterization of E. bieneusi in hamsters is available worldwide. In this study, fecal specimens were collected from 175 golden hamsters and 175 Siberian hamsters purchased from pet shops in three provinces of China. The average infection rate of E. bieneusi was 12.0% (42/350), with 14.9% (26/175) in pet golden hamsters and 9.1% (16/175) in pet Siberian hamsters. Four genotypes were identified in pet golden hamsters, including three known genotypes (D, Henan-II, and SHW5) and one novel genotype (named Ebph1). Five genotypes were found in pet Siberian hamsters, including one known genotype (D) and four novel genotypes (named Ebph2 to Ebph5). Genotypes D and Ebph2 were the dominant genotype in pet golden hamsters (23/26, 88.5%) and Siberian hamsters (9/16, 56.3%), respectively. Phylogenetic analysis showed that the E. bieneusi isolates clustered into two groups: Group 1 (D, Henan-II, SHW5, and Ebph1) and Group 3 (Ebph2 to Ebph5). To the best of our knowledge, this is the first report of E. bieneusi infection in golden hamsters and Siberian hamsters worldwide. The identification of four genotypes belonging to Group 1 of high zoonotic potential suggests that pet hamsters especially golden hamsters can be potential sources of human microsporidiosis.
Résumé
Enterocytozoon bieneusi, un agent pathogène opportuniste commun, a été détecté chez les humains et un large éventail d’animaux dans le monde. Cependant, aucune information sur la prévalence et la caractérisation moléculaire d’E. bieneusi chez les hamsters n’est disponible. Dans cette étude, des échantillons fécaux ont été prélevés sur 175 hamsters dorés et 175 hamsters sibériens achetés dans des animaleries de trois provinces de Chine. Le taux d’infection moyen d’E. bieneusi était de 12,0 % (42/350), avec 14,9 % (26/175) chez les hamsters dorés et 9,1 % (16/175) chez les hamsters sibériens. Quatre génotypes ont été identifiés chez les hamsters dorés, dont trois génotypes connus (D, Henan-II et SHW5) et un nouveau génotype (nommé Ebph1). Cinq génotypes ont été trouvés chez des hamsters sibériens, dont un génotype connu (D) et quatre nouveaux génotypes (nommés Ebph2 à Ebph5). Les génotypes D et Ebph2 étaient les génotypes dominants, respectivement chez les hamsters dorés (23/26, 88,5 %) et les hamsters sibériens (9/16, 56,3 %). L’analyse phylogénétique a montré que les isolats d’E. bieneusi se regroupaient en deux groupes : le groupe 1 (D, Henan-II, SHW5 et Ebph1) et le groupe 3 (Ebph2 à Ebph5). À notre connaissance, il s’agit du premier signalement d’infection par E. bieneusi chez des hamsters dorés et des hamsters de Sibérie dans le monde. L’identification de quatre génotypes appartenant au groupe 1, à fort potentiel zoonotique, suggère que les hamsters de compagnie, en particulier les hamsters dorés, peuvent être des sources potentielles de microsporidiose humaine.
Key words: Enterocytozoon bieneusi / Golden hamsters / Siberian hamsters / Genotype / Zoonotic / China
© C. Lv et al., published by EDP Sciences, 2022
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Enterocytozoon bieneusi is a common microsporidian parasite that infects humans and a broad range of mammals and birds [14]. The parasite can cause asymptomatic infection or self-limiting diarrhea in immunocompetent hosts, and life-threatening diarrhea in immunocompromized individuals such as human immunodeficiency virus (HIV)-infected patients, organ transplant recipients, and cancer patients [7]. Hosts acquire infection usually through fecal–oral transmission of spores from infected humans and animals via direct contact or by ingestion of spore-contaminated food or water [14].
Sequence analysis based on the internal transcribed spacer (ITS) region of the rRNA gene has identified 11 major phylogenetic groups and more than 500 genotypes of E. bieneusi from humans and animals [14]. Groups 1 and 2, which are of major public health importance, contain most potential zoonotic genotypes; while other groups (Groups 3–11) appear to be more host-specific, with apparently limited public health importance [14]. Nearly 30 rodent species belonging to nine families (Castoridae, Caviidae, Chinchillidae, Cricetidae, Echimyidae, Hystricidae, Muridae, Sciuridae, and Spalacidae) have been reported as hosts of E. bieneusi, and approximately 100 E. bieneusi genotypes have been identified in rodents worldwide [4–6, 8, 11, 13, 16, 18, 19, 25–27, 30–33]. Among them, genotype D is the most common detected from rodents in previous studies [24, 27].
Hamsters are extremely popular small pets that are kept by both young children and adults worldwide. In China, the most common pet hamster species are golden hamster (Mesocricetus auratus), and two dwarf hamsters, namely Siberian hamster (Phodopus sungorus) and Campbell hamster (Phodopus campbelli). Pet rodents can carry a variety of zoonotic pathogens including viruses, bacteria, and parasites (such as Cryptosporidium, Giardia, and E. bieneusi) [17, 20, 27]; zoonotic transmission of E. bieneusi has occurred from domestic guinea pigs to a child in Peru [3]. However, no information is available on the occurrence and genetic characteristics of E. bieneusi in hamsters worldwide. The purpose of the present study was to determine the prevalence and zoonotic potential of E. bieneusi in these animals.
Materials and methods
Ethics statement
The research protocol was reviewed and approved by the Research Ethics Committee of Henan University of Science and Technology.
Sample collection
From September 2018 to October 2019, a total of 350 fecal specimens were collected from the two most common species (n = 175 each) of pet hamsters purchased from seven pet shops in three cities, i.e., Luoyang (Henan Province), Weifang (Shandong Province), and Xuzhou (Jiangsu Province) in China (Table 1). The hamsters in pet shops were all pets offered for sale. From each pet shop, 25 golden hamsters (Mesocricetus auratus) and 25 Siberian hamsters (Phodopus sungorus) were obtained. Upon arrival at the laboratory, each animal was immediately placed into a separate clean plastic box for collection of fresh feces. Each animal was raised separately, and only a single sample was collected from each animal. All the specimens were stored at 4 °C prior to DNA extraction (within one week). Only 1–10-month-old pet hamsters were available in these pet shops. All pet hamsters examined in this study showed no clinical signs of disease at the time of sample collection, and information on region, age, and gender of these animals was recorded.
Prevalence and genotypes of Enterocytozoon bieneusi in pet golden hamsters (Mesocricetus auratus) and Siberian hamsters (Phodopus sungorus) in China.
DNA extraction
Each fecal specimen was washed with distilled water by centrifugation at 3000 r·min−1 for 10 min. Genomic DNA was extracted from approximately 200 mg processed fecal samples using an E.Z.N.A. Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA), according to the procedure recommended by the manufacturer. The extracted DNA was stored at −20 °C before it was used for PCR amplification.
Molecular detection
DNA from each specimen was tested for the presence of E. bieneusi by nested PCR targeting a ~390-bp fragment of the ITS region, as previously described [2]. The external primers were EBITS3 (5′–GGTCATAGGGATGAAGAG–3′) and EBITS4 (5′–TTCGAGTTCTTTCGCGCTC–3′), whereas the internal primers were EBITS1 (5′–GCTCTGAATATCTATGGCT–3′) and EBITS2.4 (5′–ATCGCCGACGGATCCAAGTG–3′). 2×EasyTaq® PCR SuperMix (TransGen Biotech, Beijing, China) were used for PCR amplification. The thermal-cycling procedure was used as previously described [27]. Positive control (DNA of guinea pig-derived genotype S7) and negative control (distilled water) were included in each PCR analysis. Secondary PCR products were examined by agarose gel electrophoresis and visualized by staining with GelStain (TransGen Biotech, Beijing, China).
Nucleotide sequencing and phylogenetic analysis
Two-directional sequencing of positive PCR products was done by General Biol (Anhui, China). The nucleotide sequences obtained were aligned with available sequences in GenBank, using ClustalX 2.1 (http://www.clustal.org/) and MegAlign, version 7.1 (a tool in the software DNAStar, https://www.dnastar.com/). Genotypes of E. bieneusi were determined and named based on ~243 bp of the ITS region, according to the established nomenclature system [21]. A neighbor-joining tree was generated using MEGA 7 software (http://www.megasoftware.net/). The evolutionary distances were computed using the maximum composite likelihood method, and the reliability of branches in the tree was assessed by bootstrap analysis using 1000 replicates.
Statistical analysis
Chi-square analysis was performed to assess the correlation between the prevalence of E. bieneusi and the age, gender, and region of pet golden hamsters and Siberian hamsters using SPSS, version 17.0 (Statistical Package for the Social Sciences). A difference was considered statistically significant when the p value was <0.05.
Nucleotide sequence accession numbers
Representative ITS nucleotide sequences of E. bieneusi obtained from pet golden hamsters and Siberian hamsters in this study have been deposited in GenBank under accession numbers OM427481–OM427489.
Results and discussion
To our knowledge, this is the first report of E. bieneusi infection in pet hamsters. In the present study, 26 (14.9%) of 175 pet golden hamsters and 16 (9.1%) of 175 pet Siberian hamsters were positive for E. bieneusi by PCR, with an average infection rate of 12.0%. Compared with other pet rodents, the prevalence of E. bieneusi in pet hamsters was higher than pet chinchillas (3.6%), similar to pet fancy rats (11.2%), but lower than pet squirrels (16.7%), chipmunks (17.6%), and guinea pigs (20.2%) [5, 6, 19, 27]. The infection rates of E. bieneusi in pet rodents could be influenced by many factors, such as animal species, host health status, age distribution, management and living conditions, different geographical regions, and sample sizes.
The prevalence of E. bieneusi in pet golden hamsters was higher than that in pet Siberian hamsters, but the difference was not significant (p > 0.05). In both golden hamsters and Siberian hamsters, although the prevalences of E. bieneusi in younger animals were higher than those in older ones, the differences in the prevalence of E. bieneusi in both species between different region, age and gender groups were not significant (p > 0.05) (Table 1). This finding was in accordance with the observations in previous studies on pet red-bellied tree squirrels, fancy rats and guinea pigs in China [5, 27].
Eight E. bieneusi genotypes, including three known genotypes (D, Henan-II, and SHW5) and five novel genotypes (named Ebph1–Ebph5), were identified by ITS sequence analysis in this study (Table 1). Genotype Ebph1 differed from D by two nucleotides. Ebph2 differed from WL4 (AY237212) by one nucleotide. Genotypes Ebph3 to Ebph5 differed from WL4 (AY237212) by two nucleotides each. Sequence differences among the five novel genotypes in the present study and their relatives (D or WL4) are shown in Figure 1. In the phylogenetic tree of the E. bieneusi ITS region, genotypes D, Henan-II, SHW5, and Ebph1 clustered into Group 1 of high zoonotic potential [13], and four novel genotypes Ebph2–Ebph5 clustered within the strong host-specific Group 3 (Fig. 2).
 |
Fig. 1 Nucleotide sequence diversity at the ITS region of Enterocytozoon bieneusi genotypes obtained in this study in comparison with related reference sequences. |
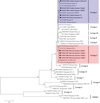 |
Fig. 2 Phylogenetic relationships among the genotypes of E. bieneusi identified in this study and other known genotypes, as inferred by a neighbor-joining analysis of the ITS region. Bootstrap values above 50% from 1000 pseudoreplicates are shown. The genotypes identified in this study are indicated by closed circles. |
The E. bieneusi genotype distributions in the two hamster species were apparently different. In pet golden hamsters, genotype D (n = 23, 88.5%) was the predominant genotype, followed by Henan-II, SHW5, and Ebph1 (n = 1 each) (Table 1). For pet Siberian hamsters, the dominant genotype was Ebph2 (n = 9, 56.3%), followed by D (n = 4, 25.0%), and Ebph3–Ebph5 (n = 1 each) (Table 1). All the four genotypes in pet golden hamsters belonged to zoonotic Group 1; while expect for genotype D, the other four genotypes in pet Siberian hamsters were divided into host-specific Group 3. The difference is probably due to different host species. However, this observation is based on limited sampling sizes and regions, further studies on more samples collected from different regions should be conducted to understand the genetic diversity of E. bieneusi from hamsters in China.
In the present study, genotype D was the dominant genotype in pet golden hamsters, which is consistent with several previous reports from rodents such as pet and wild rats, pet red-bellied tree squirrels, pet red squirrels, domestic bamboo rats, and wild mice [4, 5, 13, 26, 27]. Genotype D is one of the most common zoonotic genotypes worldwide, and has been reported in several human cases in China [14, 29]. Genotype D has a broad range of animal hosts in China, such as non-human primates, livestock, companion animals, wildlife, and birds, as well as in wastewater [13, 14, 27]. In the present study, genotype D was identified in hamsters for the first time.
The genotype Henan-II was previously reported in an HIV-positive patient in Henan, China [28], and was found in a pet golden hamster for the first time. Genotype SHW5 was recently identified in wastewater samples from a hospital in Shanghai, China [10], and was found in a pet golden hamster in this study for the first time. Therefore, pet golden hamsters can be the source of human infection with the two genotypes, and more studies are needed to understand their host range and public health importance.
Six Group 3 genotypes have previously been reported, including WL6, WL22, WL23, WL25, PtEb VIII, and WL4 [13]. The first four genotypes are restricted to four rodent species in the USA [9, 23], and PtEb VIII has been found only in a cat in Portugal [15]. In contrast, genotype WL4 has been reported in a wider range of animal hosts, including wild rodents (squirrels, chipmunks, deer mice, and muskrats), and other mammals (raccoons, bears, otters, ermines, deer, and cottontails) in the USA [9, 22, 23], as well as in water samples in the USA, Tunisia, and China [1, 9, 12]. In the present study, the four novel genotypes (Ebph2–Ebph5) identified in pet Siberian hamsters, differed from WL4 by 1–2 nucleotides and clustered into Group 3. Genotypes Ebph2–Ebph5 may have a narrow host range and low or no public health importance.
Conclusions
This is the first report of E. bieneusi infection in pet golden hamsters and Siberian hamsters. Three known genotypes and five novel genotypes were identified in this study, with zoonotic genotype D and host-specific genotype Ebph2 being the dominant genotype in pet golden hamsters and Siberian hamsters, respectively. The identification of four genotypes belonging to zoonotic Group 1 suggests that pet hamsters may be potential sources of E. bieneusi infection in humans. Therefore, pet owners, especially children, should be educated to take precautions to reduce the transmission risk.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
This study was supported by the Young Academic Leader Training Project of Henan University of Science and Technology (13490009), Student Research Training Program (SRTP) in Henan University of Science and Technology (2019392) and Henan Province (S201910464052).
References
- Ben Ayed L, Yang W, Widmer G, Cama V, Ortega Y, Xiao L. 2012. Survey and genetic characterization of wastewater in Tunisia for Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. Journal of Water and Health, 10, 431–444. [CrossRef] [PubMed] [Google Scholar]
- Buckholt MA, Lee JH, Tzipori S. 2002. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Applied and Environmental Microbiology, 68, 2595–2599. [CrossRef] [PubMed] [Google Scholar]
- Cama VA, Pearson J, Cabrera L, Pacheco L, Gilman R, Meyer S, Ortega Y, Xiao L. 2007. Transmission of Enterocytozoon bieneusi between a child and guinea pigs. Journal of Clinical Microbiology, 45, 2708–2710. [CrossRef] [PubMed] [Google Scholar]
- Deng L, Chai Y, Luo R, Yang L, Yao J, Zhong Z, Wang W, Xiang L, Fu H, Liu H, Zhou Z, Yue C, Chen W, Peng G. 2020. Occurrence and genetic characteristics of Cryptosporidium spp. and Enterocytozoon bieneusi in pet red squirrels (Sciurus vulgaris) in China. Scientific Reports, 10, 1026. [CrossRef] [PubMed] [Google Scholar]
- Deng L, Li W, Yu X, Gong C, Liu X, Zhong Z, Xie N, Lei S, Yu J, Fu H, Chen H, Xu H, Hu Y, Peng G. 2016. First report of the human-pathogenic Enterocytozoon bieneusi from red-bellied tree squirrels (Callosciurus erythraeus) in Sichuan, China. PLoS One, 11, e0163605. [CrossRef] [PubMed] [Google Scholar]
- Deng L, Li W, Zhong Z, Chai Y, Yang L, Zheng H, Wang W, Fu H, He M, Huang X, Zuo Z, Wang Y, Cao S, Liu H, Ma X, Wu K, Peng G. 2018. Molecular characterization and new genotypes of Enterocytozoon bieneusi in pet chipmunks (Eutamias asiaticus) in Sichuan province, China. BMC Microbiology, 18, 37. [CrossRef] [PubMed] [Google Scholar]
- Didier ES, Weiss LM. 2006. Microsporidiosis: current status. Current Opinion in Infectious Diseases, 19, 485–492. [CrossRef] [PubMed] [Google Scholar]
- Gui BZ, Zou Y, Chen YW, Li F, Jin YC, Liu MT, Yi JN, Zheng WB, Liu GH. 2020. Novel genotypes and multilocus genotypes of Enterocytozoon bieneusi in two wild rat species in China: potential for zoonotic transmission. Parasitology Research, 119, 283–290. [CrossRef] [PubMed] [Google Scholar]
- Guo Y, Alderisio KA, Yang W, Cama V, Feng Y, Xiao L. 2014. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Applied and Environmental Microbiology, 80, 218–225. [CrossRef] [PubMed] [Google Scholar]
- Jiang W, Roellig DM, Li N, Wang L, Guo Y, Feng Y, Xiao L. 2020. Contribution of hospitals to the occurrence of enteric protists in urban wastewater. Parasitology Research, 119, 3033–3040. [CrossRef] [PubMed] [Google Scholar]
- Li J, Jiang Y, Wang W, Chao L, Jia Y, Yuan Y, Wang J, Qiu J, Qi M. 2020. Molecular identification and genotyping of Enterocytozoon bieneusi in experimental rats in China. Experimental Parasitology, 210, 107850. [Google Scholar]
- Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, Guo M, Liu L, Feng Y. 2012. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Neglected Tropical Diseases, 6, e1809. [CrossRef] [PubMed] [Google Scholar]
- Li W, Feng Y, Santin M. 2019. Host specificity of Enterocytozoon bieneusi and public health implications. Trends in Parasitology, 35, 436–451. [CrossRef] [PubMed] [Google Scholar]
- Li W, Xiao L. 2021. Ecological and public health significance of Enterocytozoon bieneusi. One Health, 12, 100209. [CrossRef] [PubMed] [Google Scholar]
- Lobo ML, Xiao L, Cama V, Stevens T, Antunes F, Matos O. 2006. Genotypes of Enterocytozoon bieneusi in mammals in Portugal. Journal of Eukaryotic Microbiology, 53(Suppl 1), S61–S64. [Google Scholar]
- Masuda A, Wada M, Saho H, Tokunaga K, Kikuchi Y, Yamasaki F, Matsumoto J. 2021. Prevalence and molecular characterization of the zoonotic enteric protozoans Cryptosporidium spp., Enterocytozoon bieneusi, and Blastocystis from pallas’s squirrels (Callosciurus erythraeus) in Kanagawa Prefecture, Japan. Microbiology Spectrum, 9, e0099021. [CrossRef] [PubMed] [Google Scholar]
- Meerburg BG, Singleton GR, Kijlstra A. 2009. Rodent-borne diseases and their risks for public health. Critical Reviews in Microbiology, 35, 221–270. [CrossRef] [PubMed] [Google Scholar]
- Ni HB, Sun YZ, Qin SY, Wang YC, Zhao Q, Sun ZY, Zhang M, Yang D, Feng ZH, Guan ZH, Qiu HY, Wang HX, Xue NY, Sun HT. 2021. Molecular detection of Cryptosporidium spp. and Enterocytozoon bieneusi infection in wild rodents from six provinces in China. Frontiers in Cellular and Infection Microbiology, 11, 783508. [CrossRef] [PubMed] [Google Scholar]
- Qi M, Luo N, Wang H, Yu F, Wang R, Huang J, Zhang L. 2015. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitology International, 64, 339–341. [CrossRef] [PubMed] [Google Scholar]
- Ryan UM, Feng Y, Fayer R, Xiao L. 2021. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia - a 50 year perspective (1971–2021). International Journal for Parasitology, 51, 1099–1119. [CrossRef] [PubMed] [Google Scholar]
- Santin M, Fayer R. 2009. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. Journal of Eukaryotic Microbiology, 56, 34–38. [CrossRef] [Google Scholar]
- Santin M, Fayer R. 2015. Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. Journal of Eukaryotic Microbiology, 62, 34–43. [CrossRef] [Google Scholar]
- Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW 3rd, Xiao L. 2003. Molecular characterization of microsporidia indicates that wild mammals Harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Applied and Environmental Microbiology, 69, 4495–4501. [CrossRef] [PubMed] [Google Scholar]
- Taghipour A, Bahadory S, Abdoli A, Javanmard E. 2021. A systematic review and meta-analysis on the global molecular epidemiology of microsporidia infection among rodents: a serious threat to public health. Acta Parasitologica. https://doi.org/10.1007/s11686-021-00447-8. [PubMed] [Google Scholar]
- Tavalla M, Kazemi F, Mardani-Kateki M, Abdizadeh R. 2018. Molecular diagnosis of Enterocytozoon bieneusi and Encephalitozoon spp. in wild rats of southwest of Iran. Jundishapur Journal of Microbiology, 11, e55961. [Google Scholar]
- Wang H, Liu Q, Jiang X, Zhang Y, Zhao A, Cui Z, Li D, Qi M, Zhang L. 2019. Dominance of zoonotic genotype D of Enterocytozoon bieneusi in bamboo rats (Rhizomys sinensis). Infection, Genetics and Evolution, 73, 113–118. [CrossRef] [PubMed] [Google Scholar]
- Wang J, Lv C, Zhao D, Zhu R, Li C, Qian W. 2020. First detection and genotyping of Enterocytozoon bieneusi in pet fancy rats (Rattus norvegicus) and guinea pigs (Cavia porcellus) in China. Parasite, 27, 21. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, Liu L, Feng Y, Xiao L. 2013. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. Journal of Clinical Microbiology, 51, 557–563. [CrossRef] [PubMed] [Google Scholar]
- Wang SS, Wang RJ, Fan XC, Liu TL, Zhang LX, Zhao GH. 2018. Prevalence and genotypes of Enterocytozoon bieneusi in China. Acta Tropica, 183, 142–152. [Google Scholar]
- Xu J, Wang X, Jing H, Cao S, Zhang X, Jiang Y, Yin J, Cao J, Shen Y. 2020. Identification and genotyping of Enterocytozoon bieneusi in wild Himalayan marmots (Marmota himalayana) and Alashan ground squirrels (Spermophilus alashanicus) in the Qinghai-Tibetan Plateau area (QTPA) of Gansu Province, China. Parasites & Vectors, 13, 367. [CrossRef] [PubMed] [Google Scholar]
- Yu F, Cao Y, Wang H, Liu Q, Zhao A, Qi M, Zhang L. 2020. Host-adaptation of the rare Enterocytozoon bieneusi genotype CHN4 in Myocastor coypus (Rodentia: Echimyidae) in China. Parasites & Vectors, 13, 578. [CrossRef] [PubMed] [Google Scholar]
- Yu F, Qi M, Zhao Z, Lv C, Wang Y, Wang R, Zhang L. 2019. The Potential role of synanthropic rodents and flies in the transmission of Enterocytozoon bieneusi on a dairy cattle farm in China. Journal of Eukaryotic Microbiology, 66, 435–441. [CrossRef] [PubMed] [Google Scholar]
- Zhao W, Wang J, Ren G, Yang Z, Yang F, Zhang W, Xu Y, Liu A, Ling H. 2018. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasites & Vectors, 11, 313. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Lv C, Wang J, Li C, Zhang M & Qian W. 2022. First detection and genotyping of Enterocytozoon bieneusi in pet golden hamsters (Mesocricetus auratus) and Siberian hamsters (Phodopus sungorus) in China. Parasite 29, 15.
All Tables
Prevalence and genotypes of Enterocytozoon bieneusi in pet golden hamsters (Mesocricetus auratus) and Siberian hamsters (Phodopus sungorus) in China.
All Figures
 |
Fig. 1 Nucleotide sequence diversity at the ITS region of Enterocytozoon bieneusi genotypes obtained in this study in comparison with related reference sequences. |
| In the text | |
 |
Fig. 2 Phylogenetic relationships among the genotypes of E. bieneusi identified in this study and other known genotypes, as inferred by a neighbor-joining analysis of the ITS region. Bootstrap values above 50% from 1000 pseudoreplicates are shown. The genotypes identified in this study are indicated by closed circles. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


