| Issue |
Parasite
Volume 26, 2019
|
|
|---|---|---|
| Article Number | 62 | |
| Number of page(s) | 7 | |
| DOI | https://doi.org/10.1051/parasite/2019064 | |
| Published online | 31 October 2019 | |
Research Article
Genotype identification and phylogenetic analysis of Enterocytozoon bieneusi in farmed black goats (Capra hircus) from China’s Hainan Province
Identification du génotype et analyse phylogénétique d’Enterocytozoon bieneusi chez des chèvres noires (Capra hircus) de la province de Hainan, en Chine
1
Department of Pathogenic Biology, Hainan Medical University, Xueyuan Road 3, 571199 Haikou, Hainan, PR China
2
Hainan Medical University-The University of Hong Kong Joint Laboratory of Tropical Infectious Diseases, Hainan Medical University, 571199 Haikou, Hainan, PR China
3
Key Laboratory of Tropical Translational Medicine of Ministry of Education, Hainan Medical University, 571199 Haikou, PR China
4
Institute of Animal Science and Veterinary Medicine, Hainan Academy of Agricultural Sciences, 571100 Haikou, PR China
5
College of Animal Sciences, Tarim University, 843300 Alar, Xinjiang, PR China
* Corresponding authors: luganghn@163.com;
** hayidazhaowei@163.com
Received:
6
September
2019
Accepted:
16
October
2019
Enterocytozoon bieneusi is an important pathogen commonly found in humans and animals. Farmed animals with close contact to humans are important hosts of E. bieneusi. The role of goats in the transmission of E. bieneusi, however, remains unclear. In this study, 341 fresh fecal samples of black goats were collected from five locations in Hainan Province, China. Enterocytozoon bieneusi was identified and genotyped by sequences of the internal transcribed spacer (ITS) region. Phylogenetic analysis was performed by constructing a neighbor-joining tree of the ITS gene sequences. The average prevalence of E. bieneusi in black goats was 24.0% (82/341) with rates ranging from 6.3% (4/63) to 37.2% (32/86) across the locations (χ2 = 17.252, p < 0.01). Eight genotypes of E. bieneusi were identified, including six known genotypes: CHG5 (n = 47); CHG3 (n = 23); CHG2 (n = 4); CM21 (n = 3); D (n = 2); and AHG1 (n = 1), and two novel genotypes termed HNG-I (n = 1) and HNG-II (n = 1). In the phylogenetic tree, genotype D was clustered into Group 1 and the other identified genotypes were included in Group 2. This represents the first report identifying E. bieneusi in black goats from Hainan Province, with a high prevalence and wide occurrence demonstrated. The two new genotypes identified provide additional insights into the genotypic variations in E. bieneusi. Due to the small percentage of zoonotic genotypes in these animals, there is minimal risk of zoonotic transmission of E. bieneusi.
Résumé
Enterocytozoon bieneusi est un agent pathogène important que l’on trouve couramment chez l’homme et les animaux. Les animaux d’élevage, en contact étroit avec l’homme, sont des hôtes importants d’E. bieneusi. Le rôle des chèvres dans la transmission d’E. bieneusi reste toutefois incertain. Dans cette étude, 341 échantillons de fèces fraîches de chèvres noires ont été prélevés dans cinq sites de la province de Hainan, en Chine. Enterocytozoon bieneusi a été identifié et génotypé par des séquences de la région de l’espaceur interne transcrit (ITS). L’analyse phylogénétique a été réalisée en construisant un arbre de jonction voisine des séquences du gène ITS. La prévalence moyenne d’E. bieneusi chez les chèvres noires était de 24,0 % (82/341), avec des taux allant de 6,3 % (4/63) à 37,2 % (32/86) dans tous les sites (χ2 = 17,252, p < 0,01). Huit génotypes d’E. bieneusi ont été identifiés, dont six génotypes connus: CHG5 (n = 47) ; CHG3 (n = 23) ; CHG2 (n = 4) ; CM21 (n = 3) ; D (n = 2) ; AHG1 (n = 1) et deux nouveaux génotypes appelés HNG-I (n = 1) et HNG-II (n = 1). Dans l’arbre phylogénétique, le génotype D appartenait au groupe 1 et les autres génotypes identifiés étaient inclus dans le groupe 2. Il s’agit du premier rapport identifiant E. bieneusi chez des chèvres noires de la province de Hainan, avec une prévalence élevée et une occurrence étendue. Les deux nouveaux génotypes identifiés fournissent des informations supplémentaires sur les variations génotypiques chez E. bieneusi. En raison du faible pourcentage de génotypes zoonotiques chez ces animaux, le risque de transmission zoonotique d’E. bieneusi est minime.
Key words: Enterocytozoon bieneusi / Genotype / ITS region / Goats / China
© H-H. Zhou et al., published by EDP Sciences, 2019
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Enterocytozoon bieneusi is one of the most common species of microsporidia contributing to human microsporidiosis, which generally causes digestive disorders, including diarrhea, particularly in the young and in adults with immunodeficiency [17]. E. bieneusi is frequently found in numerous animal hosts worldwide, raising public health concerns regarding its zoonotic transmission [20]. The primary mode of infection by E. bieneusi is the fecal-oral route. E. bieneusi spores can be acquired from infected humans or animals through contaminated food and water [6]. Tracing the sources of contamination and elucidating the transmission routes of E. bieneusi represent important steps to control E. bieneusi infection in humans.
Sequence analysis of the ribosomal internal transcribed spacer (ITS) region is the standard method for the detection and identification of E. bieneusi genotypes [19]. To date, more than 500 genotypes have been identified, of which 130 have been found in humans [7, 13, 15, 26]. The known genotypes of E. bieneusi were divided into 11 phylogenetic groups (Groups 1–11) by phylogenetic analysis. Up to 90% of human pathogenic genotypes belong to Group 1 or Group 2, and the genotypes within these groups have been reported in a range of diverse hosts, highlighting the zoonotic nature of the disease and cross-species infections [13]. However, the genotypes in Groups 3–11 appear more amenable to host adaptation [13]. The contribution of each animal source to human infections is poorly understood. This can be clarified by the genotyping of E. bieneusi in different animals.
The goat industry plays a dominant role in animal husbandry in China. In recent years, a number of studies of E. bieneusi have been performed in goats, particularly those in China [1, 3, 4, 14, 16, 18, 21, 22, 28]. A total of 46 ITS genotypes of E. bieneusi have been identified in goats, 11 of which have been detected in humans [1, 3, 4, 13, 14, 16, 18, 21, 22, 28]. Nevertheless, the genotypes of E. bieneusi in goats in China are not fully understood.
Hainan black goats are native to Hainan, China and represent a common breed due to their tolerance to local hot and wet weather. They are the main goat breed in Hainan, China, where the goat industry represents an important source of poverty alleviation [8]. These animals are often in close contact with keepers, and their feces are commonly excreted directly into the surrounding environment without treatment. Environmental contamination with E. bieneusi spores represents a threat to public health, but no information on E. bieneusi in these animals is available. The aims of this study were to examine the prevalence and genotyping of E. bieneusi from farmed black goats in Hainan Province, the southernmost region of China.
Materials and methods
Collection of fecal specimens
A total of 341 fresh fecal specimens from black goats were collected from Ding’an (n = 52), Chengmai (n = 63), Wenchang (n = 63), Ledong (n = 77), and Wanning (n = 86) in Hainan Province from September 2018 to March 2019 (Fig. 1). Sampled goats belonged to two groups: the first consisting of 73 animals aged ≤ 3 months (kids) and the other group consisting of 268 animals aged ≥ 4 months (older goats). The farms were selected based only on the owners’ willingness to participate and the accessibility of animals for sampling. All the goats were maintained in pens on the farms. Fecal samples were collected from pens housing 3–6 goats. To avoid the chance of duplicate sampling of animals, only one fecal specimen was collected in each pen. All the fecal specimens were collected at the bed boards immediately after defecation by using a sterile disposable latex glove and then placed in labeled sterile tubes individually. All bags were transported to our laboratory in a cooler with ice packs (<24 h) and stored at −20 °C until processing (<1 w).
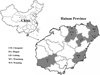 |
Figure 1 Specific locations where samples were collected in this study. ● Sampling points. |
DNA extraction
All fecal specimens were sieved through an 8.0-cm-diameter sieve with a pore size of 45 μm, and filtrates were concentrated by centrifugation at 1500 g for 10 min. DNA was extracted from ~200 mg of each processed fecal specimen using a QIAamp DNA stool mini kit (QIAgen, Hilden, Germany). Extracted DNA was stored at −20 °C prior to PCR analysis.
PCR amplification
Nested PCRs were performed to amplify a ~390-bp region containing 76 bp of the 3″ end of the SSU rRNA gene, 243 bp of the internal gene, and 70 bp of the 5′ region of the large-subunit (LSU) rRNA gene. The aim was to detect the presence of E. bieneusi in all the extracted DNA samples using two pairs of primers, EBITS3 and EBITS4, and EBITS1 and EBITS2.4 for the first and the second amplifications, respectively. The primers and cycling parameters were designed by Buckholt et al., as follows: the outer primers were EBITS3 (5′–GGTCATAGGGATGAAGAG–3′) and EBITS4 (5′–TTCGAGTTCTTTCGCGCTC–3′) and the inner primers were EBITS1 (5′–GCTCTGAATATCTATGGCT–3′) and EBITS2.4 (5′–ATCGCCGACGGATCCAAGTG–3′). The two sets of cycling parameters were 35 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 40 s and 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 40 s, with both of them having a final extension step at 72 °C for 10 min. These reactions produced fragments of 435 and 390 bp, respectively [2]. TaKaRa Taq DNA Polymerase (TaKaRa Bio Inc., Tokyo, Japan) was used for all PCR amplifications. A negative control with no DNA was amplified in all PCR tests, including the amplification of negative controls from the first PCR in the second PCR reaction, to ensure low levels of contamination. All secondary PCR products were analyzed on 1.5% agarose gels and visualized by GelRed staining (Biotium Inc., Hayward, CA, USA).
Nucleotide sequencing and analysis
All secondary PCR products positive for E. bieneusi were sent for sequencing (Sangon Biotech Co., Ltd., Shanghai, China) and all were sequenced in both directions. The nucleotide sequences obtained were aligned with each other and reference sequences downloaded from the GenBank database using Clustal X 1.83 (http://www.clustal.org/) to determine the genotypes of E. bieneusi isolates. The genotypes of E. bieneusi obtained in this study were given the first published name when identical to known genotypes in GenBank. Meanwhile, the genotypes that produced ITS sequences with single nucleotide substitutions, deletions, or insertions confirmed by DNA sequencing of at least two PCR products were considered novel genotypes. All were given a genotype name through the addition of roman numbers behind the abbreviation HNG (Hainan Goat), according to their order of appearance. All genotypes were named based on a 243 bp region of the ITS gene of E. bieneusi according to the established nomenclature system [20].
Phylogenetic analysis
To confirm the genogroup designation and to assess the genetic relationships of novel ITS genotypes of E. bieneusi obtained, phylogenetic analysis was performed by constructing a neighbor-joining tree using the program Mega X (http://www.megasoftware.net/) based on the evolutionary distances calculated by the Kimura-2-parameter model. The reliability of these trees was assessed using bootstrap analysis with 1000 replicates.
Statistical analysis
Data entry and analysis were performed using Social Sciences (SPSS) 19.0 software. The statistical significance of differences in infection proportions was generally evaluated by Pearson’s Chi-square test. The significant level of all tests was: p-value = 0.05.
Nucleotide sequence accession numbers
Representative nucleotide sequences obtained in the study were deposited in the GenBank database under accession numbers MN267058 and MN267059.
Results
Occurrence of E. bieneusi in black goats
A total of 24.0% (82/341) of black goat samples were positive for E. bieneusi by PCR and sequencing analysis. E. bieneusi was identified in all five locations with the highest infection rate of 37.2% (32/86) in Wanning, followed by 36.5% (19/52) in Ding’an, 30.2% (19/63) in Chengmai, 10.4% (8/77) in Ledong, and the lowest infection rate of 6.3% (4/63) in Wenchang. There were significant differences in prevalence among these locations (χ2 = 17.252, p < 0.01). The prevalence rates were 38.4% (28/73) and 20.1% (54/268) in kids and older goats, respectively, suggesting significant differences between them (χ2 = 10.413, p < 0.01) (Table 1).
Prevalence and genotype distribution of E. bieneusi isolates from goats farmed in different cities in Hainan Province.
Genetic characterization and genotypic distribution of E. bieneusi in black goats
Sequence analysis demonstrated that the 82 E. bieneusi isolates belonged to eight ITS genotypes including six known genotypes (AHG1, CHG2, CHG3, CHG5, CM21 and D), and two novel genotypes (HNG-I and HNG-II). There were 34 polymorphic sites among the eight genotypes identified (Fig. 2). The novel genotypes HNG-I (MN267058) and HNG-II (MN267059) had the largest similarity with genotype CHG5 (KP262365) with 10 and with one base difference, respectively. Among the genotypes, CHG5 (57.3%, 47/82) dominated, followed by CHG3 (28.0%, 23/82), CHG2 (4.9%, 4/82), CM21 (3.7%, 3/82), D (2.4%, 2/82), and each of the remaining three genotypes AHG1, HNG-I and HNG-II (1.2%, 1/82) (Table 1).
 |
Figure 2 Sequence variation in the ITS region of the rRNA gene of E. bieneusi isolates identified in black goats. Dots indicate the same base identity as the ITS gene sequence of genotype CHG5. |
The distributions of E. bieneusi genotypes in animals according to location and age are shown in Table 1. Genotypes D, AHG1 and HNG-II were only found in Wenchang, while genotypes HNG-I and CHG2 were only present in Ding’an and Wanning, respectively. However, genotypes CHG3, CHG5 and CM21 were found in four, three and two areas, respectively. Regarding the age groups, genotypes CHG2, CHG3, CHG5, and CM21 were found in two age groups, while genotypes D, HNG-I and HNG-II were only found in older goats; genotype AHG1 was only found in kids.
Phylogenetic relationships of E. bieneusi genotypes
Based on the phylogenetic analysis of the neighbor-joining tree of the ITS gene sequences of E. bieneusi, all identified genotypes with the exception of genotype D, were in Group 2 (Fig. 3).
 |
Figure 3 Phylogenetic tree based on the neighbor-joining analysis of ITS sequences. The phylogenetic relationships of E. bieneusi genotypes identified here and other known genotypes deposited in GenBank were inferred by a neighbor-joining analysis of ITS sequences based on genetic distance by the Kimura 2-parameter model. The numbers on the branches are percent bootstrapping values from 1000 replicates. Each sequence is identified by its accession number, host origin, and genotype designation. The E. bieneusi genotype CSK2 (KY706128) from white kangaroo was used as the outgroup. The squares and triangles filled in black indicate novel and known genotypes identified in this study, respectively. |
Discussion
To date, there have been nine reports on the identification and genotyping of E. bieneusi in goats [1, 3, 4, 14, 16, 18, 21, 22, 28]. In this study, the prevalence of E. bieneusi in black goats was 24.0% (82/341) which was lower than that in Chongqing (62.5%; 5/8) [21] and Henan (31.2%; 186/596) [14, 18, 21], but higher than that in Anhui (5.1%; 33/654) [14, 21], Yunnan (12.8%; 60/470) [4, 21], Jiangsu (2.7%; 2/74) [14], Heilongjiang (21.8%; 12/55) [28], Shaanxi (19.9%; 72/361) [18, 21] and Tibet (9.6%; 25/260) [3]. Elsewhere, infection rates of 14.3% (1/7), 13.3% (11/83), and 19.2% (14/73) have been reported in goats from Spain [16], Egypt [1] and Thailand [22], respectively. The results suggested that goats are important hosts of E. bieneusi and may thus play a key role in the transmission of microsporidiosis caused by E. bieneusi.
In terms of age groups, we observed a significantly higher infection rate of E. bieneusi in kids (38.4%; 28/73) than in older goats (20.1%; 54/268) in Hainan (χ2 = 10.413, p < 0.01), which was consistent with previous findings [18, 21]. A recent study showed that the prevalence of E. bieneusi decreased with increasing age [4]. The high risk of younger goats to infection with E. bieneusi can be explained through their lower immune status and higher disease susceptibility.
To date, 46 genotypes of E. bieneusi have been identified in goats worldwide, 11 of which have been detected in humans [1, 3, 4, 14, 16, 18, 21, 22, 28]. In this study, eight genotypes were identified in goats, including six known genotypes (AHG1, CHG2, CHG3, CHG5, CM21 and D), and two novel genotypes (HNG-I and HNG-II). Out of the eight genotypes, genotype CHG5 was the most prevalent, with a prevalence of 57.3% (47/82), followed by CHG3 which had a prevalence of 28.0% (23/82) and then the remaining six genotypes with a lower prevalence of 14.6% (12/82). These results were different from those observed in other provinces of China. For example, the dominant genotypes found in Tibet were EbpC and EbpA, in Shaanxi genotype SX1, in Henan and Yunnan genotype BEB6, and in Anhui genotype CHG3. Therefore, E. bieneusi infections in goats likely differ by region.
All the studies on E. bieneusi in goats found at least one zoonotic genotype (BEB6, Peru6, EbpA and EbpC) with a high prevalence, except for the study from Anhui Province. Interestingly, Peru6 and EbpC were commonly found in humans from Yunnan and Heilongjiang Provinces; they were also found in animals including pigs, minks and birds [5, 7, 15, 27, 29]. Zoonotic genotype BEB6 was found in a child from Shanghai City, and it seems more common in herbivore animals in China, including deer, sheep, goats, cattle and horses [30]. These findings suggest that goats can transmit the above-mentioned zoonotic genotypes to humans and other animals. However, we did not identify genotypes BEB6, Peru6, EbpA and EbpC, but we found genotype D, which is responsible for the majority of human infections and has been found in humans in more than 40 countries or areas [13, 17]. In China, this genotype has been found in AIDS patients, cancer patients, children, and HIV-positive patients from Henan, Shanghai, Hubei, and Heilongjiang Provinces, summarized by Li et al. [13]. It has also been reported as the dominant genotype in no-human primates, horses, pigs, and cats, as well as in urban wastewater [1, 9–11, 23, 24, 27]. These results illustrate that the interspecies transmission of genotype D poses a high zoonotic risk, representing a public health concern within the human population.
The other five known genotypes (AHG1, CHG2, CHG3, CHG5 and CM21) identified were found in sheep or goats in previous studies [14, 18, 21]. Genotypes CHG2 and CHG3 have also been detected in dairy cattle [4, 12], and genotype CM21 was found in captive golden snub-nosed monkeys [25]. However, genotypes CHG5 and AHG1 were only found in goats [14, 18, 21]. To date, the potential of five known genotypes identified and the two novel genotypes (HNG-I and HNG-II) to cause disease in humans or other livestock is unknown. Their host adaptation and potential role in the zoonotic transmission of E. bieneusi infection now requires further study in more systematic molecular epidemiological investigations of E. bieneusi in a larger number of hosts.
Conclusion
In conclusion, this is the first report on the identification of E. bieneusi from farmed black goats in Hainan Province, with high prevalence and wide occurrence demonstrated. It is necessary to develop improved farm management to prevent the occurrence of cross-transmission and re-infection of E. bieneusi between different individuals within each goat farm. Although the six known genotypes identified here have already been reported in goats, two novel genotypes were identified and provide novel insights into the genotypic variation in E. bieneusi. However, due to the small percentage of potentially zoonotic genotypes in these animals, there is a minimal risk of zoonotic transmission of E. bieneusi.
Acknowledgments
This work was supported by the Innovation Research Team Project of Hainan Natural Science Foundation (2018CXTD340) and the National Natural Science Foundation of China (Nos. 81672072 and 81760378). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Competing interests
The authors declare that they have no competing interests.
References
- Al-Herrawy AZ, Gad MA. 2016. Microsporidial spores in fecal samples of some domesticated animals living in Giza, Egypt. Iran Journal of Parasitology, 11(2), 195–203. [Google Scholar]
- Buckholt MA, Lee JH, Tzipori S. 2002. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Applied & Environmental Microbiology, 68(5), 2595–2599. [CrossRef] [PubMed] [Google Scholar]
- Chang Y, Wang Y, Wu Y, Niu MZ, Li J, Zhang S, Wang R, Jian F, Ning C, Zhang L. 2019. Molecular characterization of Giardia duodenalis and Enterocytozoon bieneusi isolated from Tibetan sheep and Tibetan goats under natural grazing conditions in Tibet. Journal of Eukaryotic Microbiology. DOI: 10.1111/jeu.12758. [Epub ahead of print]. [Google Scholar]
- Chen D, Wang SS, Zou Y, Li Z, Xie SC, Shi LQ, Zou FC, Zhu XQ, Yang JF, Zhao GH. 2018. Prevalence and multi-locus genotypes of Enterocytozoon bieneusi in black-boned sheep and goats in Yunnan Province, southwestern China. Infection, Genetics and Evolution, 65, 385–391. [CrossRef] [Google Scholar]
- Cong W, Qin SY, Meng QF. 2018. Molecular characterization and new genotypes of Enterocytozoon bieneusi in minks (Neovison vison) in China. Parasite, 25, 34. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Fayer R, Santin-Duran M. 2014. Epidemiology of microsporidia in human infections, in Microsporidia: pathogens of opportunity, Weiss LM, Becnel JJ, Editors, first ed. John Wiley & Sons, Inc: Chichester, UK. DOI: 10.1002/9781118395264.ch3. [Google Scholar]
- Gong B, Yang Y, Liu X, Cao J, Xu M, Xu N, Yang F, Wu F, Li B, Liu A, Shen Y. 2019. First survey of Enterocytozoon bieneusi and dominant genotype Peru6 among ethnic minority groups in southwestern China’s Yunnan Province and assessment of risk factors. PLoS Neglected Tropical Diseases, 13, e0007356. [CrossRef] [PubMed] [Google Scholar]
- Hu J, Zhao W, Niu L, Wang L, Li L, Zhang H, Zhong T. 2016. Gene organization and characterization of the complete mitochondrial genome of Hainan black goat (Capra hircus). Mitochondrial DNA Part A, DNA Mapping, Sequencing, and Analysis, 27, 1656–1657. [PubMed] [Google Scholar]
- Karim MR, Dong H, Yu F, Jian F, Zhang L, Wang R, Zhang S, Rume FI, Ning C, Xiao L. 2014. Genetic diversity in Enterocytozoon bieneusi isolates from dogs and cats in China: host specificity and public health implications. Journal of Clinical Microbiology, 52, 3297–3302. [CrossRef] [PubMed] [Google Scholar]
- Karim MR, Wang R, Dong H, Zhang L, Li J, Zhang S, Rume FI, Qi M, Jian F, Sun M, Yang G, Zou F, Ning C, Xiao L. 2014. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Applied & Environmental Microbiology, 80, 1893–1898. [Google Scholar]
- Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, Guo M, Liu L, Feng Y. 2012. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Neglected Tropical Diseases, 6(9), e1809. [CrossRef] [PubMed] [Google Scholar]
- Li J, Luo N, Wang C, Qi M, Cao J, Cui Z, Huang J, Wang R, Zhang L. 2016. Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasites & Vectors, 9, 142. [CrossRef] [PubMed] [Google Scholar]
- Li W, Feng Y, Santin M. 2019. Host specificity of Enterocytozoon bieneusi and public health implications. Trends in Parasitology, 35(6), 436–451. [CrossRef] [PubMed] [Google Scholar]
- Li WC, Wang K, Gu YF. 2019. Detection and genotyping study of Enterocytozoon bieneusi in sheep and goats in East-central China. Acta Parasitology, 64(1), 44–50. [CrossRef] [Google Scholar]
- Liu H, Jiang Z, Yuan Z, Yin J, Wang Z, Yu B, Zhou D, Shen Y, Cao J. 2017. Infection by and genotype characteristics of Enterocytozoon bieneusi in HIV/AIDS patients from Guangxi Zhuang autonomous region, China. BMC Infectious Diseases, 17(1), 684. [CrossRef] [PubMed] [Google Scholar]
- Lores B, del Aguila C, Arias C. 2002. Enterocytozoon bieneusi (microsporidia) in faecal samples from domestic animals from Galicia, Spain. Memorias do Instituto Oswaldo Cruz, 97(7), 941–945. [CrossRef] [PubMed] [Google Scholar]
- Matos O, Lobo ML, Xiao L. 2012. Epidemiology of Enterocytozoon bieneusi infection in humans. Journal of Parasitology Research, 2012, 981424. [CrossRef] [PubMed] [Google Scholar]
- Peng XQ, Tian GR, Ren GJ, Yu ZQ, Lok JB, Zhang LX, Wang XT, Song JK, Zhao GH. 2016. Infection rate of Giardia duodenalis, Cryptosporidium spp. and Enterocytozoon bieneusi in cashmere, dairy and meat goats in China. Infection, Genetics and Evolution, 41, 26–31. [CrossRef] [Google Scholar]
- Santín M, Fayer R. 2009. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. Journal of Eukaryotic Microbiology, 56(1), 34–38. [CrossRef] [Google Scholar]
- Santín M, Fayer R. 2011. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Research in Veterinary Science, 90(3), 363–371. [CrossRef] [PubMed] [Google Scholar]
- Shi K, Li M, Wang X, Li J, Karim MR, Wang R, Zhang L, Jian F, Ning C. 2016. Molecular survey of Enterocytozoon bieneusi in sheep and goats in China. Parasites & Vectors, 9, 23. [CrossRef] [PubMed] [Google Scholar]
- Udonsom R, Prasertbun R, Mahittikorn A, Chiabchalard R, Sutthikornchai C, Palasuwan A, Popruk S. 2019. Identification of Enterocytozoon bieneusi in goats and cattle in Thailand. BMC Veterinary Research, 15(1), 308. [CrossRef] [PubMed] [Google Scholar]
- Wagnerová P, Sak B, Květoňová D, Buňatová Z, Civišová H, Maršálek M, Kváč M. 2012. Enterocytozoon bieneusi and Encephalitozoon cuniculi in horses kept under different management systems in the Czech Republic. Veterinary Parasitology, 190(3–4), 573–577. [CrossRef] [PubMed] [Google Scholar]
- Wang SS, Li JQ, Li YH, Wang XW, Fan XC, Liu X, Li ZJ, Song JK, Zhang LX, Zhao GH. 2018. Novel genotypes and multilocus genotypes of Enterocytozoon bieneusi in pigs in northwestern China: a public health concern. Infection, Genetics and Evolution, 63, 89–94. [CrossRef] [Google Scholar]
- Yu F, Wu Y, Li T, Cao J, Wang J, Hu S, Zhu H, Zhang S, Wang R, Ning C, Zhang L. 2017. High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Veterinary Research, 13, 158. [CrossRef] [PubMed] [Google Scholar]
- Zhang Y, Koehler AV, Wang T, Robertson GJ, Bradbury RS, Gasser RB. 2018. Enterocytozoon bieneusi genotypes in people with gastrointestinal disorders in Queensland and Western Australia. Infection, Genetics and Evolution, 65, 293–299. [CrossRef] [Google Scholar]
- Zhao W, Zhang W, Yang F, Cao J, Liu H, Yang D, Shen Y, Liu A. 2014. High prevalence of Enterocytozoon bieneusi in asymptomatic pigs and assessment of zoonotic risk at the genotype level. Applied & Environmental Microbiology, 80(12), 3699–3707. [CrossRef] [PubMed] [Google Scholar]
- Zhao W, Zhang W, Yang D, Zhang L, Wang R, Liu A. 2015. Prevalence of Enterocytozoon bieneusi and genetic diversity of ITS genotypes in sheep and goats in China. Infection, Genetics and Evolution, 32, 265–270. [CrossRef] [Google Scholar]
- Zhao W, Yu S, Yang Z, Zhang Y, Zhang L, Wang R, Zhang W, Yang F, Liu A. 2016. Genotyping of Enterocytozoon bieneusi (Microsporidia) isolated from various birds in China. Infection, Genetics and Evolution, 40, 151–154. [CrossRef] [Google Scholar]
- Zhao W, Wang J, Yang Z, Liu A. 2017. Dominance of the Enterocytozoon bieneusi genotype BEB6 in red deer (Cervus elaphus) and Siberian roe deer (Capreolus pygargus) in China and a brief literature review. Parasite, 24, 54. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
Cite this article as: Zhou H-H, Zheng X-L, Ma T-M, Qi M, Cao Z-X, Chao Z, Wei L-M, Liu Q-W, Sun R-P, Wang F, Zhang Y, Lu G & Zhao W. 2019. Genotype identification and phylogenetic analysis of Enterocytozoon bieneusi in farmed black goats (Capra hircus) from China’s Hainan Province. Parasite 26, 62.
All Tables
Prevalence and genotype distribution of E. bieneusi isolates from goats farmed in different cities in Hainan Province.
All Figures
 |
Figure 1 Specific locations where samples were collected in this study. ● Sampling points. |
| In the text | |
 |
Figure 2 Sequence variation in the ITS region of the rRNA gene of E. bieneusi isolates identified in black goats. Dots indicate the same base identity as the ITS gene sequence of genotype CHG5. |
| In the text | |
 |
Figure 3 Phylogenetic tree based on the neighbor-joining analysis of ITS sequences. The phylogenetic relationships of E. bieneusi genotypes identified here and other known genotypes deposited in GenBank were inferred by a neighbor-joining analysis of ITS sequences based on genetic distance by the Kimura 2-parameter model. The numbers on the branches are percent bootstrapping values from 1000 replicates. Each sequence is identified by its accession number, host origin, and genotype designation. The E. bieneusi genotype CSK2 (KY706128) from white kangaroo was used as the outgroup. The squares and triangles filled in black indicate novel and known genotypes identified in this study, respectively. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


