Figure 1.
Download original image
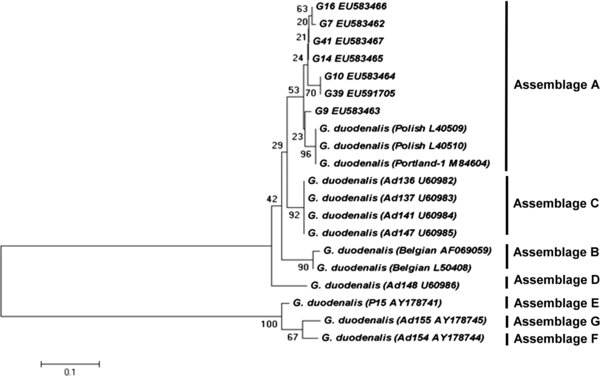
Phylogenetic relationship between the 7 Giardia Assemblages (G7, G9, G10, G14, G16, G41 and G39) obtained in this study and other previously published Giardia Assemblages. Nucleotide sequences of the GDH gene obtained in this study were aligned against reference sequences retrieved from GenBank using ClustalW and MEGA 5. The evolutionary history was inferred using the Neighbor-Joining method and the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method with the pairwise deletion option and are in the units of the number of base substitutions per site.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


