| Issue |
Parasite
Volume 27, 2020
|
|
|---|---|---|
| Article Number | 1 | |
| Number of page(s) | 7 | |
| DOI | https://doi.org/10.1051/parasite/2019078 | |
| Published online | 07 January 2020 | |
Research Article
Prevalence and morphological and molecular characteristics of Sarcocystis bertrami in horses in China
Prévalence et caractéristiques morphologiques et moléculaires de Sarcocystis bertrami chez les chevaux en Chine
1
School of Biological Sciences, Yunnan University, Kunming 650091, PR China
2
Southeast Asia Biodiversity Research Institute, Chinese Academy of Science, Yezin, Nay Pyi Taw 05282, Myanmar
3
College of Veterinary Medicine, Yangzhou University, Yangzhou 225009, PR China
4
Electron Microscope Laboratory, Kunming Medical University, Kunming 650500, PR China
* Corresponding author: jjhu@ynu.edu.cn
Received:
10
July
2019
Accepted:
20
December
2019
Three cyst-forming Sarcocystis species have been identified in horsemeat; however, there exists considerable confusion concerning their relationships. Here, 74% (34/46) of the examined tissue samples from horses contained sarcocysts based on examination by light microscopy (LM), and the organism was identified as Sarcocystis bertrami based on cyst structure. The S. bertrami cysts were microscopic (up to 6750 μm in length) and exhibited a striated wall with 2.0–5.1 μm villar protrusions (vps) under LM. Transmission electron microscopy (TEM) observations showed that the vps were tightly packed, similar to “type 11c”. Four genetic markers (18S, 28S, ITS1 and the mitochondrial cox1 gene) of S. bertrami were sequenced and analyzed. The 28S and ITS1 sequences are the first records for Sarcocystis in horses. The newly obtained sequences of the 18S and cox1 genes both shared the highest similarities with those of S. bertrami and S. fayeri obtained from horses. Phylogenetic analysis based on the 18S, 28S and cox1 sequences revealed that S. bertrami and S. fayeri formed an independent clade within a group comprising Sarcocystis spp. from ruminants and pigs. Therefore, S. bertrami and S. fayeri are considered to represent the same species of Sarcocystis in horses, and S. fayeri is a junior synonym of Sarcocystis bertrami.
Résumé
Trois espèces de Sarcocystis formant des kystes ont été identifiées dans la viande de cheval, mais il existe une confusion considérable concernant leurs relations. Ici, 74 % (34/46) des échantillons de tissus examinés provenant de chevaux contenaient des sarcocystes selon l’examen par microscopie optique (LM), et l’organisme a été identifié comme Sarcocystis bertrami selon la structure du kyste. Les kystes de S. bertrami étaient microscopiques (jusqu’à 6 750 μm de longueur) et présentaient une paroi striée avec des saillies villaires (vp) de 2,0 à 5,1 μm sous LM. Les observations en microscopie électronique à transmission ont montré que les vp étaient très serrées, similaires au « type 11c ». Quatre marqueurs génétiques (18S, 28S, ITS1 et le gène mitochondrial cox1) de S. bertrami ont été séquencés et analysés. Les séquences de 28S et ITS1 sont les premières de Sarcocystis chez les chevaux. Les séquences nouvellement obtenues de 18S et des gènes mitochondriaux cox1 partagent les similitudes les plus élevées avec celles de S. bertrami et S. fayeri obtenues à partir de chevaux. Une analyse phylogénétique basée sur les séquences de 18S, 28S et cox1 a révélé que S. bertrami et S. fayeri formaient un clade indépendant au sein d’un groupe comprenant les Sarcocystis spp. des ruminants et des porcs. Par conséquent, S. bertrami et S. fayeri sont considérés comme représentant la même espèce de Sarcocystis chez les chevaux, et S. fayeri est un synonyme plus récent de Sarcocystis bertrami.
Key words: Horse / Sarcocystis bertrami / Prevalence / Systematics / Morphology
© C.-L. Ma et al., published by EDP Sciences, 2020
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Four Sarcocystis species have been named in horses (Equus caballus) so far: S. bertrami Doflein, 1901 [1]; S. equicanis Rommel and Geidel, 1975 [22]; S. fayeri Dubey, Streitel, Stromberg, and Toussant, 1977 [2]; and S. neurona Dubey, Davis, Speer, Bowman, de Lahunta, Granstrom, Topper, Hamir, Cummings, and Suter, 1991 [3]. Mature sarcocysts of S. bertrami, S. equicanis and S. fayeri have been found in horses, and all of these species have dogs as their definitive hosts. The horse is considered an aberrant host of S. neurona because only schizonts have been identified in horses with certainty. There is currently considerable confusion concerning the validity of the three abovementioned cyst-forming Sarcocystis species in horses and other equids [4].
Traditionally, sarcocyst structure and life cycle are the two major criteria for naming a new species of Sarcocystis in a given intermediate host. However, the morphological characteristics of sarcocysts in horses have been observed to undergo some changes in various stages of development [6, 17]. In the past decade, molecular analysis based on nucleotide sequences has been recommended as a useful and efficient tool for delineating or identifying species of Sarcocystis from the same or different hosts [10–12, 14, 15]. The 18S rDNA and mitochondrial cox1 sequences of S. bertrami and S. fayeri have recently been sequenced and deposited in GenBank. However, the relationship of the two species inferred from molecular data is still unclear owing to research work performed and published by different research teams almost simultaneously [19, 23]. There are currently no records of the 28S rDNA and ITS-1 sequences of Sarcocystis spp. from horses available in GenBank.
In the present study, the prevalence of Sarcocystis species in horsemeat in China was investigated based on the morphological characteristics of the sarcocysts. Additionally, four genetic markers, 18S rDNA, 28S rDNA, ITS-1 and mitochondrial cox1 of the parasite were sequenced and analyzed to augment its descriptions and explore the relationship with Sarcocystis spp. in horses.
Materials and methods
Morphological examination of sarcocysts from horses
The study protocol was approved by the Animal Ethics Committee of Yunnan University (permission number: AECYU2015021). Horsemeat serves as a food source for humans and is commonly marketed in China. In total, tissues from 46 horses were examined from two abattoirs, one in Kunming City and another in Shilin Prefecture, both of which are located in Yunnan Province, China, from October 2015 to June 2016. From each animal, fresh tissue samples (50 g each) from the esophagus, diaphragm, skeletal muscle, tongue, and heart were examined for sarcocysts. In the laboratory, 40 specimens of approximately 10 × 3 mm in size from each collected sample were pressed and squeezed between two glass slides and then inspected using stereomicroscopy. Thereafter, individual sarcocysts were extracted and isolated from skeletal muscular fibers using needles and processed for light microscopy (LM), transmission electron microscopy (TEM) and DNA analysis.
For TEM, four sarcocysts (two from horse No. 3 and two from horse No. 8) were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4) at 4 °C, postfixed in 1.0% osmium tetroxide in the same buffer, dehydrated in a graded alcohol series, and embedded in an Epon-Araldite mixture. Ultrathin sections were stained with uranyl acetate and lead citrate and then examined using a JEM100-CX transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 100 kV.
DNA isolation, PCR amplification, cloning, and sequence analysis
Five individual sarcocysts (cysts 3, 8, 10, 15 and 26) obtained from muscle samples from five horses (Nos. 3, 8, 10, 15 and 26) were subjected to genomic DNA extraction using the phenol/chloroform method after 0.01% proteinase K and 0.25% trypsin digestion. 18S rDNA was amplified with the S1/S4 primers [7]; 28S rDNA was amplified with the KL1/KL3, KL4/KL5b and KL6a/KL2 primers sets [18]; mitochondrial cox1 was amplified with the SF1/SR9 primers [10, 11]; and ITS-1 was amplified with the SU1F/5.8SR2 primer pair [12]. The PCR products were purified, cloned, sequenced, and characterized using the methods detailed in a previous paper [14].
Results
Observations of sarcocysts of S. bertrami by LM and TEM
Only sarcocysts resembling those of S. bertrami were found in 34 of 46 horses (73.9%). The highest prevalence was 63.0% (29/46), recorded in the esophagus, followed by 43.5% (20/46) in skeletal muscle, 23.9% (11/46) in the diaphragm, and 15.2% (7/46) in tongue muscle, whereas none were found in the heart.
Under LM, S. bertrami sarcocysts were observed to be microscopic, measuring 950–6754 × 65–130 μm (n = 30). The sarcocyst wall exhibited numerous 2.0–5.1 μm (n = 40)-long villar protrusions (vps) (Fig. 1A). The sarcocysts were septate, and their interior compartments were filled with bradyzoites measuring 14.3–16.1 × 3.1–4.8 μm (n = 30) in size.
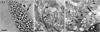 |
Figure 1 Morphological characteristics of Sarcocystis bertrami sarcocysts isolated from the skeletal muscle of a horse. (A) Sarcocyst (unstained, light microscopy) bound by villar protrusions (vps). Scale bar = 10 μm. (B) Longitudinal section of a sarcocyst (under transmission electron microscopy, TEM). The sarcocyst is surrounded by the host cell (hc), and the sarcocyst wall exhibits numerous vps, which are often bent along the cyst surface. The vps contain bundled microtubules (mt) in the core, which penetrate diagonally into the ground substance (gs). Scale bar = 1 μm. (C) Cross-section of a sarcocyst under TEM. Note the bundle of mt in the gs. Scale bar = 1 μm. |
The ultrastructure of the sarcocysts was similar to “type 11c”. The primary sarcocyst wall exhibited numerous vps, measuring 2.0–4.4 × 0.3–0.8 μm (n = 15), which were often tightly packed and folded over the cyst surface (Fig. 1B). The vps contained bundled microtubules in the core, which penetrated diagonally into the ground substance and sometimes reached the interior border of the ground substance (Figs. 1B and 1C). The primary cyst wall presented minute undulations over the entire sarcocyst surface. There was a layer of ground substances of 0.7–1.2 μm (n = 12) in thickness immediately beneath the primary sarcocyst wall (Fig. 1C).
Molecular characterization of 18S rDNA
Genomic DNA was extracted from five individual sarcocysts, and the 18S rDNA, 28S rDNA, mitochondrial cox1 and ITS-1 sequences were amplified successfully using their DNA as templates. Four 18S rDNA nucleotide sequences for cysts 3, 8, 10 and 15, two 28S rDNA sequences for cysts 3 and 10, three mitochondrial cox1 sequences for cysts 3, 8 and 26, and five ITS-1 sequences for cysts 3, 8, 10, 15 and 26 were successfully assembled, and all of the sequences including the primers were deposited in GenBank.
The four 18S rDNA nucleotide sequences (MH025625–MH025628) were 1592–1594 bp in length and shared an identity of 99.7–99.9% (average 99.8% identity). The most similar sequences in GenBank were those of S. bertrami from horsemeat in China (KX545397–KX545404, 95.0–100% identity, average 98.0%) and S. fayeri from horsemeat from Japan, Canada, and Italy (AB661437–AB661447, AB972440–AB972443 and LC171831–LC171838, 88.9–100% identity, average 96.5% identity).
The phylogenetic tree inferred from the 18S rDNA sequences revealed that the newly obtained sequences of S. bertrami formed an individual clade with those of S. fayeri from horsemeat from Japan (AB661437), Canada (AB972443) and Italy (LC171838) within a group comprising S. miescheriana (JN256123) and S. suihominis (KP732435) from pigs using canids and primates as definitive hosts, respectively, and Sarcocystis spp. from ruminants with primates or felids as definitive hosts (Fig. 2).
 |
Figure 2 Phylogenetic tree based on 18S rDNA sequences. The tree was built using the maximum parsimony method with the Tree-Bisection-Regrafting algorithm. The analysis involved 36 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 1258 positions in the final dataset. The values between the branches represent bootstrap values per 1000 replicates, and values below 50% are not shown. The four new sequences of Sarcocystis bertrami (MH025625–MH025628, shown in boldface) formed a clade with S. fayeri from horses, and the clade was within a group comprising Sarcocystis spp. from ruminants and pigs. |
Molecular characterization of 28S rDNA
The two 28S rDNA nucleotide sequences (MH025629 and MH025630) were 3456 bp and 3449 bp in length, respectively, and shared 99.2% identity. The most similar sequence in GenBank was that of S. miescheriana (AF076902) from a pig, but the identity was only 90.4–90.5%.
The phylogenetic tree based on the 28S rDNA sequences revealed that S. bertrami formed a clade with S. miescheriana (AF076902), S. gigantea (U85706) from sheep and S. moulei (AF012884) from goats, the last two of which use felids as definitive hosts (Fig. 3).
 |
Figure 3 Phylogenetic tree based on 28S rDNA gene sequences. The tree was built using the maximum parsimony method with the Tree-Bisection-Regrafting algorithm. The analysis involved 21 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 3680 positions in the final dataset. The values between the branches represent the bootstrap values per 1000 replicates, and values below 50% are not shown. The two new sequences of Sarcocystis bertrami (MH025629 and MH025630, shown in boldface) formed a clade with S. miescheriana from a pig, and Sarcocystis spp. from ruminants. |
Molecular characterization of mitochondrial cox1
The three mitochondrial cox1 nucleotide sequences (MH025631–MH025633) were 1060–1063 bp in length and shared an identity of 98.4–99.9% (average 98.9% identity). The most similar sequences in GenBank were those of S. fayeri (LC171840–LC171857, 98.0–99.8% identity, average 99.0% identity) from horsemeat from Japan, Canada and Italy and S. bertrami from horsemeat from China (KY399751–KY399755, KY399758, KY399760–KY399762, MF152616–MF152619, 98.1–99.2% identity, average 98.8% identity).
The phylogenetic tree based on the mitochondrial cox1 sequences revealed that the newly obtained sequences of S. bertrami formed an individual clade with those of S. bertrami from horsemeat from China (KY399753) and S. fayeri from horsemeat from Japan (LC171856), Canada (LC171850) and Italy (LC171857) within a group comprising S. miescheriana (LC349978), S. suihominis (MH404228), and Sarcocystis spp. from ruminants with felids as known or suspected definitive hosts (Fig. 4).
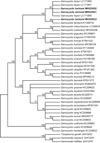 |
Figure 4 Phylogenetic tree based on mitochondrial cox1 sequences. The tree was built using the maximum parsimony method with the Tree-Bisection-Regrafting algorithm. The analysis involved 40 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 936 positions in the final dataset. The values between the branches represent the bootstrap values per 1000 replicates, and values below 50% are not shown. The three new sequences of Sarcocystis bertrami (MH025631–MH025633 shown in boldface) formed a clade with S. bertrami and S. fayeri from horses within a group comprising Sarcocystis spp. from pigs and ruminants. |
Molecular characterization of ITS-1
The five ITS-1 nucleotide sequences (MH025634–MH25638) were 934–936 bp in length and shared an identity of 96.7–98.3% (average 97.4% identity). BLAST searches using only the ITS-1 region sequences of approximately 660 bp from S. bertrami revealed that no sequences in GenBank shared significant similarities with them.
Discussion
Sarcocystis is a common parasitic protozoan with a worldwide distribution that is commonly found in a variety of mammals and birds, especially common in domesticated food animals. In the current study, the prevalence of sarcocysts in Chinese horses was 73.9% (34/46), and it has been reported to be higher than 68.8% (22/32) in Turkish horses [21], 46.2% (55/119) in Moroccan horses [16], 62.2% (245/349) in British horses [5], and 13.2% (12/91) in Belgian horses [8], but lower than 93.0% (40/43) in Mongolian horses [9].
To date, three Sarcocystis species, S. bertrami, S. equicanis and S. fayeri, have been identified from horsemeat, all of which have the dog as the definitive host. However, there is considerable confusion concerning the validity of the above species due to their similar life cycles and morphology. On the basis of TEM morphology, Dubey et al. (2016) suggested that there are two valid species of Sarcocystis in horses: a thick-walled species (S. fayeri), with a “type 11a” sarcocyst wall, and a thin-walled species (S. bertrami synonym S. equicanis), with a “type 11c” sarcocyst wall. The critical distinction between the two sarcocyst wall types is that “type 11a” exhibits nearly upright vps, but “type 11c” exhibits packed and folded vps [4]. The sarcocysts reported here were diagnosed as S. bertrami based on their similarity with “type 11c”, which has been demonstrated previously for S. equicanis from European horses [13], for Sarcocystis sp. from Mongolian horses [20] and for S. bertrami from Chinese horses [23].
Nucleotide sequence analysis has proven to be a useful tool for delineating or identifying species of Sarcocystis from the same or different hosts, and different genetic markers have shown different levels of intra- or inter-specific sequence diversity [14, 15]. There are only sequences of 18S rDNA and mitochondrial cox1 from S. bertrami and S. fayeri currently deposited in GenBank. In our analysis, the newly obtained 18S rDNA sequences exhibited up to 100% identity (average 98.0% and 96.5% identity, respectively) with those of S. bertrami and S. fayeri from horses provided in GenBank; the newly obtained mitochondrial cox1 sequences shared the highest identity of 98.0–99.8% (average 99.0%) with those of S. fayeri, followed by S. bertrami (98.1–99.2% identity, average 98.7% identity). A possible explanation for the high similarities of the two parasites is that both represent the same species of Sarcocystis in horses. The protrusions observed via TEM may appear to be upright or folded depending on the plane of the cut section. Phylogenetic analysis based on the 18S rDNA, 28S rDNA and mitochondrial cox1 sequences also confirmed the close relationship between S. bertrami and S. fayeri.
In conclusion, we found a high prevalence rate of Sarcocystis in horses in China, and only S. bertrami was identified based on the cyst ultrastructure. Based on 18S rDNA and mitochondrial cox1, S. bertrami and S. fayeri are inferred to represent the same species from horses. According to the general rules of the International Code of Zoological Nomenclature, S. fayeri should be considered a junior synonym of S. bertrami.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
This study was supported by the National Key R&D Program of China (grant 2017YFD0500400), the Natural Sciences Foundation of China (grant 31460557) and the Southeast Asia Biodiversity Research Institute, Chinese Academy of Science (grant Y4zk111B01).
References
- Doflein F. 1901. Die Protozoen als Parasiten und Krankheitserreger, nach biologischen Gesichtspunkten dargestellt. Jena XII, Germany: Gustav Fischer. p. 274 [Google Scholar]
- Dubey JP, Streitel RH, Stromberg PC, Toussant MJ. 1977. Sarcocystis fayeri sp. n. from the horse. Journal of Parasitology, 63, 443–447. [CrossRef] [Google Scholar]
- Dubey JP, Davis SW, Speer CA, Bowman DD, de Lahunta A, Granstrom DE, Topper MJ, Hamir AN, Cummings JF, Suter MM. 1991. Sarcocystis neurona n. sp. (Protozoa: Apicomplexa), the etiologic agent of equine protozoal myeloencephalitis. Journal of Parasitology, 77, 212–218. [CrossRef] [Google Scholar]
- Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R. 2016. Sarcocystosis of animals and humans, 2nd edn. Boca Raton, FL: CRC Press. p. 249–256. [Google Scholar]
- Edwards GT. 1984. Prevalence of equine Sarcocystis in British horses and a comparison of two detection methods. Veterinary Record, 115, 265–267. [CrossRef] [Google Scholar]
- Fayer R, Hounsel C, Giles RC. 1983. Chronic illness in a Sarcocystis infected pony. Veterinary Record, 113, 216–217. [CrossRef] [Google Scholar]
- Fischer S, Odening K. 1998. Characterization of bovine Sarcocystis species by analysis of their 18S ribosomal DNA sequences. Journal of Parasitology, 84, 50–54. [CrossRef] [Google Scholar]
- Fransen JLA, Degryse ADAY, van Mol KAC, Ooms LAA. 1987. Sarcocystis und chronische Myopathien bei Pferden. Berliner und Münchener tierärztliche Wochenschrift, 100, 229–232. [Google Scholar]
- Fukuyo M, Battsetseg G, Byambaa B. 2002. Prevalence of Sarcocystis infection in horses in Mongolia. Southeast Asian Journal of Tropical Medicine and Public Health, 33, 718–719. [Google Scholar]
- Gjerde B. 2013. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. International Journal for Parasitology, 43, 579–591. [CrossRef] [PubMed] [Google Scholar]
- Gjerde B. 2014. Sarcocystis species in red deer revisited: with a redescription of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology, 141, 441–452. [CrossRef] [PubMed] [Google Scholar]
- Gjerde B. 2014. Molecular characterisation of Sarcocystis rileyi from a common eider (Somateria mollissima) in Norway. Parasitology Research, 113, 3501–3509. [CrossRef] [PubMed] [Google Scholar]
- Göbel E, Rommel M. 1980. Light and electron microscopic study on cysts of Sarcocystis equicanis in the oesophageal musculature of horses. Berliner und Münchener tierärztliche Wochenschrift, 93, 41–47. [Google Scholar]
- Hu JJ, Liu TT, Liu Q, Esch GW, Chen JQ, Huang S, Wen T. 2016. Prevalence, morphology, and molecular characteristics of Sarcocystis spp. in domestic goats (Capra hircus) from Kunming, China. Parasitology Research, 115, 3973–3981. [CrossRef] [PubMed] [Google Scholar]
- Hu JJ, Huang S, Wen T, Esch GW, Liang Y, Li HL. 2017. Sarcocystis spp. in domestic sheep in Kunming City, China: prevalence, morphology, and molecular characteristics. Parasite, 4, 30. [Google Scholar]
- Kirmse P. 1986. Sarcosporidioses in equines of Morocco. British Veterinary Journal, 142, 70–72. [CrossRef] [Google Scholar]
- Matuschka FR, Schnieder T, Daugschies A, Rommel M. 1986. Cyclic transmission of Sarcocystis bertrami Doflein, 1901 by the dog to the horse. Protistologica, 22, 231–233. [Google Scholar]
- Mugridge NB, Morrison DA, Heckeroth AR, Johnson AM, Tenter AM. 1999. Phylogenetic analysis based on full-length large subunit ribosomal RNA gene sequence comparison reveals that Neospora caninum is more closely related to Hammondia heydorni than to Toxoplasma gondii. International Journal for Parasitology, 29, 1545–1556. [CrossRef] [PubMed] [Google Scholar]
- Murata R, Suzuki J, Hyuga A, Shinkai T, Sadamasu K. 2018. Molecular identification and characterization of Sarcocystis spp. in horsemeat and beef marketed in Japan. Parasite, 25, 27. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Odening K, Wesemeier HH, Walter G, Bockhardt I. 1995. Ultrastructure of sarcocysts from equids. Acta Parasitologica, 40, 12–20. [Google Scholar]
- Özer E, Saki CE, Dündar B. 1995. Tükiye’de tektirnaklilarda bulunan Sarcocystis türleri. Turkish Journal of Veterinary and Animal Sciences, 19, 177–180. [Google Scholar]
- Rommel M, Geisel O. 1975. Untersuchungen über die Verbreitung und de Lebenszyklus einer Sarkosporidienart des Pferdes (Sarcocystis equicanis n. spec.). Berliner und Münchener Tierärztliche Wochenschrif, 88, 468–471. [Google Scholar]
- Zeng W, Sun L, Xiang Z, Li N, Zhang J, He Y, Li Q, Yang F, Song J, Morris J, Rosenthal BM, Sun L, Liu H, Yang Z. 2018. Morphological and molecular characteristics of Sarcocystis bertrami from horses and donkeys in China. Veterinary Parasitology, 252, 89–94. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Ma C-L, Ye Y-L, Wen T, Huang Z-M, Pan J, Hu J-J, Tao J-P & Song J-L. 2020. Prevalence and morphological and molecular characteristics of Sarcocystis bertrami in horses in China. Parasite 27, 1.
All Figures
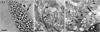 |
Figure 1 Morphological characteristics of Sarcocystis bertrami sarcocysts isolated from the skeletal muscle of a horse. (A) Sarcocyst (unstained, light microscopy) bound by villar protrusions (vps). Scale bar = 10 μm. (B) Longitudinal section of a sarcocyst (under transmission electron microscopy, TEM). The sarcocyst is surrounded by the host cell (hc), and the sarcocyst wall exhibits numerous vps, which are often bent along the cyst surface. The vps contain bundled microtubules (mt) in the core, which penetrate diagonally into the ground substance (gs). Scale bar = 1 μm. (C) Cross-section of a sarcocyst under TEM. Note the bundle of mt in the gs. Scale bar = 1 μm. |
| In the text | |
 |
Figure 2 Phylogenetic tree based on 18S rDNA sequences. The tree was built using the maximum parsimony method with the Tree-Bisection-Regrafting algorithm. The analysis involved 36 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 1258 positions in the final dataset. The values between the branches represent bootstrap values per 1000 replicates, and values below 50% are not shown. The four new sequences of Sarcocystis bertrami (MH025625–MH025628, shown in boldface) formed a clade with S. fayeri from horses, and the clade was within a group comprising Sarcocystis spp. from ruminants and pigs. |
| In the text | |
 |
Figure 3 Phylogenetic tree based on 28S rDNA gene sequences. The tree was built using the maximum parsimony method with the Tree-Bisection-Regrafting algorithm. The analysis involved 21 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 3680 positions in the final dataset. The values between the branches represent the bootstrap values per 1000 replicates, and values below 50% are not shown. The two new sequences of Sarcocystis bertrami (MH025629 and MH025630, shown in boldface) formed a clade with S. miescheriana from a pig, and Sarcocystis spp. from ruminants. |
| In the text | |
 |
Figure 4 Phylogenetic tree based on mitochondrial cox1 sequences. The tree was built using the maximum parsimony method with the Tree-Bisection-Regrafting algorithm. The analysis involved 40 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 936 positions in the final dataset. The values between the branches represent the bootstrap values per 1000 replicates, and values below 50% are not shown. The three new sequences of Sarcocystis bertrami (MH025631–MH025633 shown in boldface) formed a clade with S. bertrami and S. fayeri from horses within a group comprising Sarcocystis spp. from pigs and ruminants. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


