Fig. 2.
Download original image
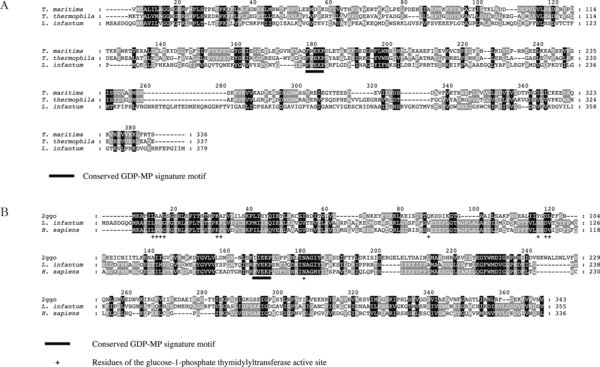
Sequence alignment of GDP-MPs.
A. Multiple sequence alignment of L. infantum, T. thermophilus, and T. maritima GDP-MPs. The L. infantum GDP-MP presents 16.1% and 15.4% of identity with the T. thermophilus and T. maritima, respectively. The signature motif F(V)EKP, essential for the activity of GDP-MPs, is underlined.
B. Multiple sequence alignment of both human and L. infantum GDP-MPs with the protein used as a template for 3D modelling (glucose-1-phosphate thymidylyltransferase from Sulfolobus tokodaii: 2ggo). This alignment shows that L. infantum and human GDP-MPs present 29% and 25% of identity with the template protein, respectively. Both human and leishmanial enzymes share 49% of identity. The amino acids of the template protein active site were identified as described in Materials & Methods. The GDP-MP signature motif F(V)EKP (underlined) is present in both human and leishmanial enzymes, but not in the template protein.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


