Figure 2
Download original image
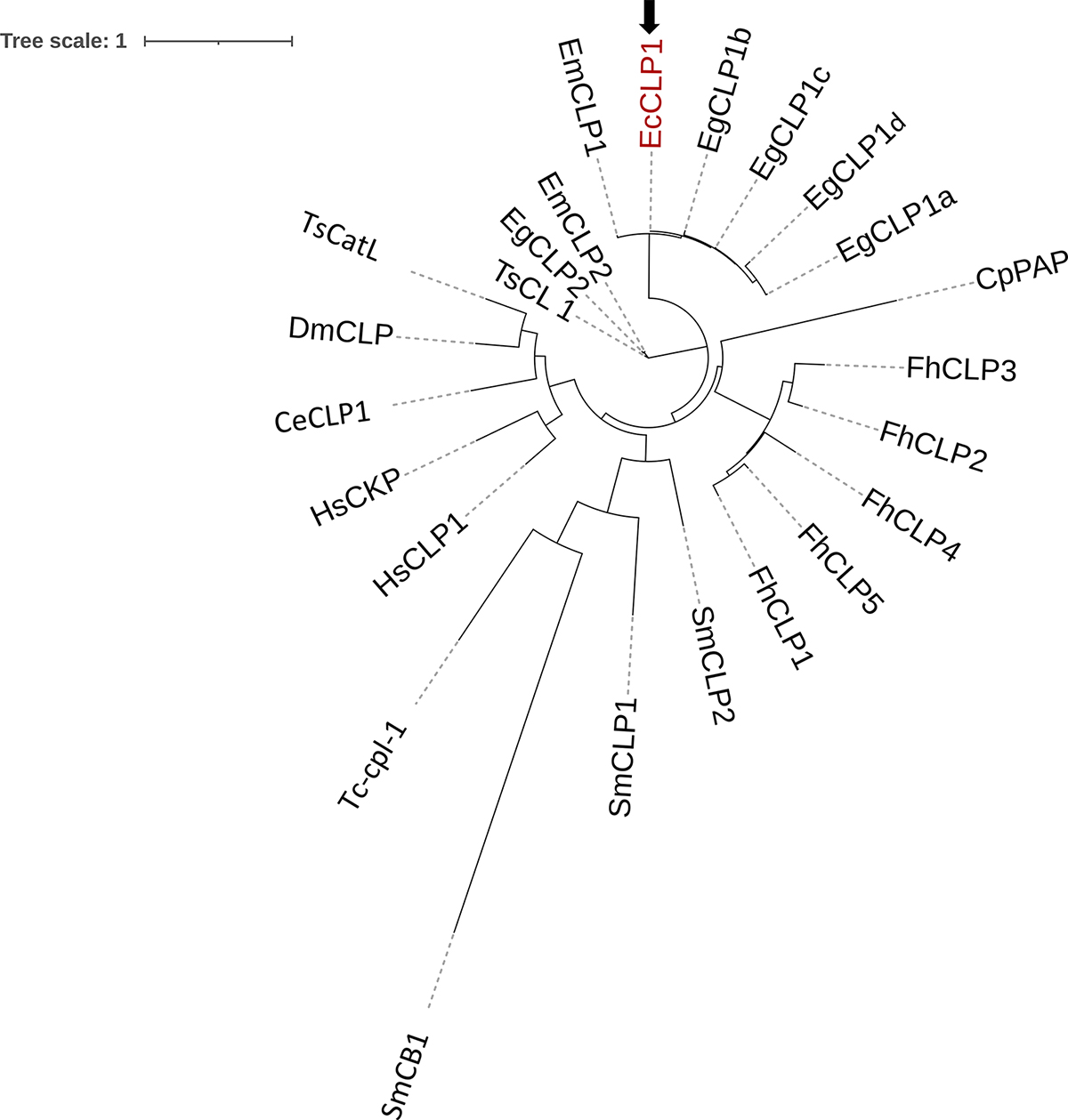
Phylogenetic tree based on the deduced amino acid sequence of EcCLP1 and cathepsin proteases from other species. The unrooted maximum likelihood tree was constructed using the phylogeny program. The black arrow indicates the EcCLP1 protease reported in this study and is marked in red letters. The accession numbers in Uniprot or GenBank databases for the species represented in the tree were the following: A0A068WJU5, A0A068WM48, A0A068WG50, A0A068WF33, U6IZB4 for the E. granulosus s.s. EgCLP1a, EgCLP1b, EgCLP1c, EgCLP1d and EgCLP2, respectively. BAF02516.1 and BAF02517.1 for the E. multilocularis EmCLP1 and EmCLP2, respectively. AAS00027.1 for the T. solium TsCL_1. AAB41670.2, AAC47721.1, QPX50259.1, ABZ80400.1 and AAF76330.1 for the F. hepatica FhCL1, FhCL2, FhCL3, FhCL4 and FhCL5, respectively. NP_001244900.1 and NP_000387.1 for the Homo sapiens HsCLP1 and HsCKP, respectively. AAB02650.1 for the papain CpPAP. O45734-1 for the C. elegans CeCLP1. AAB18345.1 for DmCLP1 or cysteine proteinase 1 from Drosophila melanogaster. AAC46485.1, Q26564_SCHMA_CLP2, and AAA29865.1 for SmCLP1, SmCLP2, and SmCB1, respectively from Schistosoma mansoni. AAC48340.1 for Tc-Cpl-1 or cathepsin L-like cysteine proteinase from Toxocara canis. KRY31298.1 for TsCatL or cathepsin L cysteine proteinase from Trichinella spiralis.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


