Figure 4
Download original image
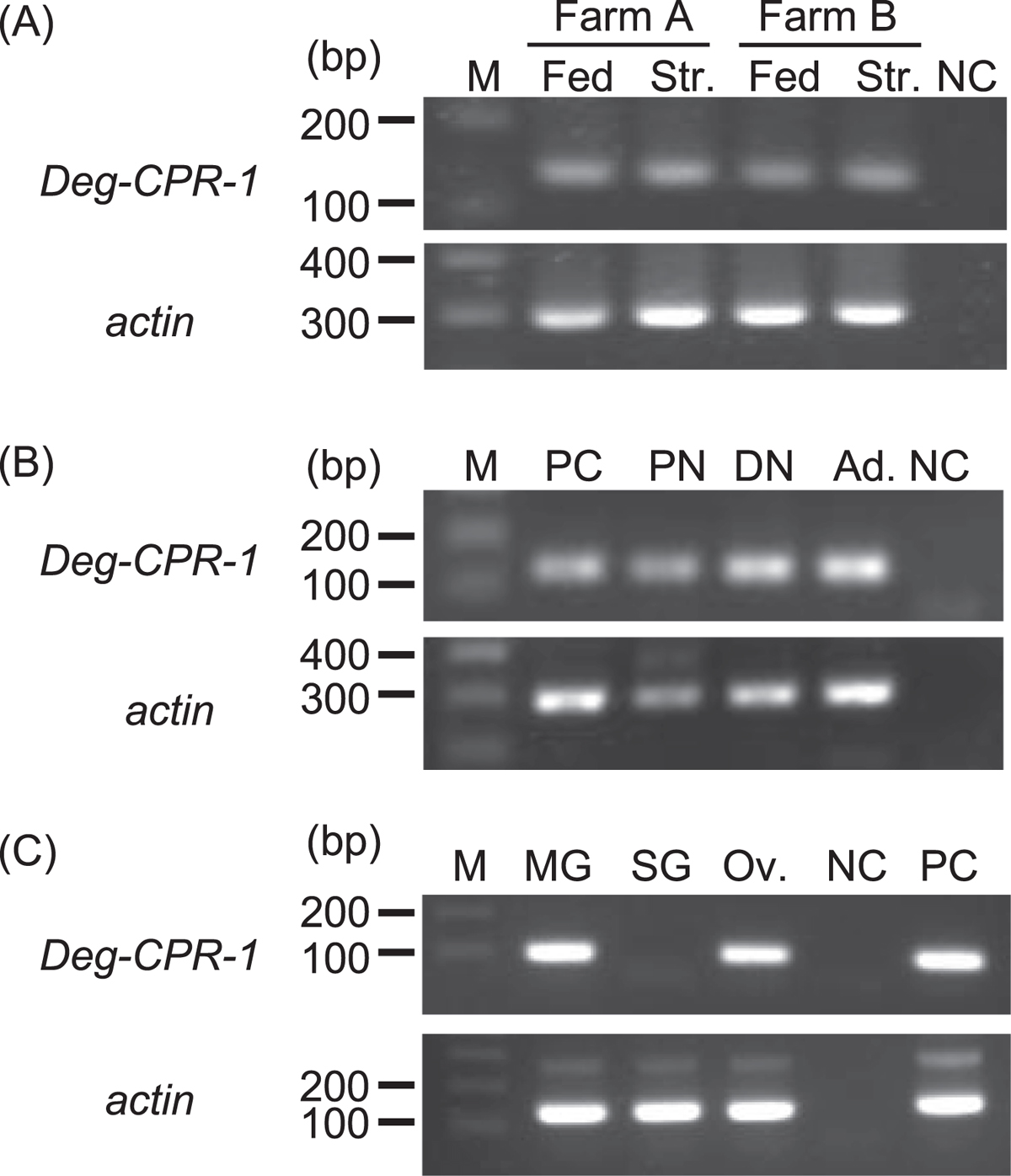
Expression analysis of the cysteine protease gene of poultry red mites (PRMs, Dermanyssus gallinae) (Deg-CPR-1). (A) Expression analysis of Deg-CPR-1 in fed and starved PRMs. Some PRMs were fixed after their transfer to the laboratory, designated as “fed PRMs”, and the remaining PRMs were maintained at 25 °C for 7 weeks to digest the blood they had ingested; these were designated as “starved PRMs”. The Deg-CPR-1 mRNA was detected in PRMs from two different farms by RT-PCR. Fed: fed PRMs, Str.: starved PRMs, NC: negative control (distilled water). (B) Expression analysis of Deg-CPR-1 in PRMs fed blood at different life stages. A portion of the starved PRMs were sorted according to their life stages, namely, protonymphs, deutonymphs, and adults, under a stereomicroscope, based on their morphology and body size, and Deg-CPR-1 mRNA was detected by RT-PCR. PC: positive control (cDNA from PRMs of all stages), PN: protonymph, DN: deutonymph, Ad: adult, NC: negative control (distilled water). (C) Expression analysis of Deg-CPR-1 in different tissues. The tissue samples were collected from starved deutonymphs and adults by laser-capture microdissection, and RT – nested PCR was performed to detect the Deg-CPR-1 mRNA in the midgut, salivary gland, and ovary. PC: positive control (cDNA from PRMs of all stages), MG: midgut, SG: salivary gland, Ov: ovary, NC: negative control (distilled water). The actin gene was amplified as an internal control in all expression analyses.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


