Figure 2
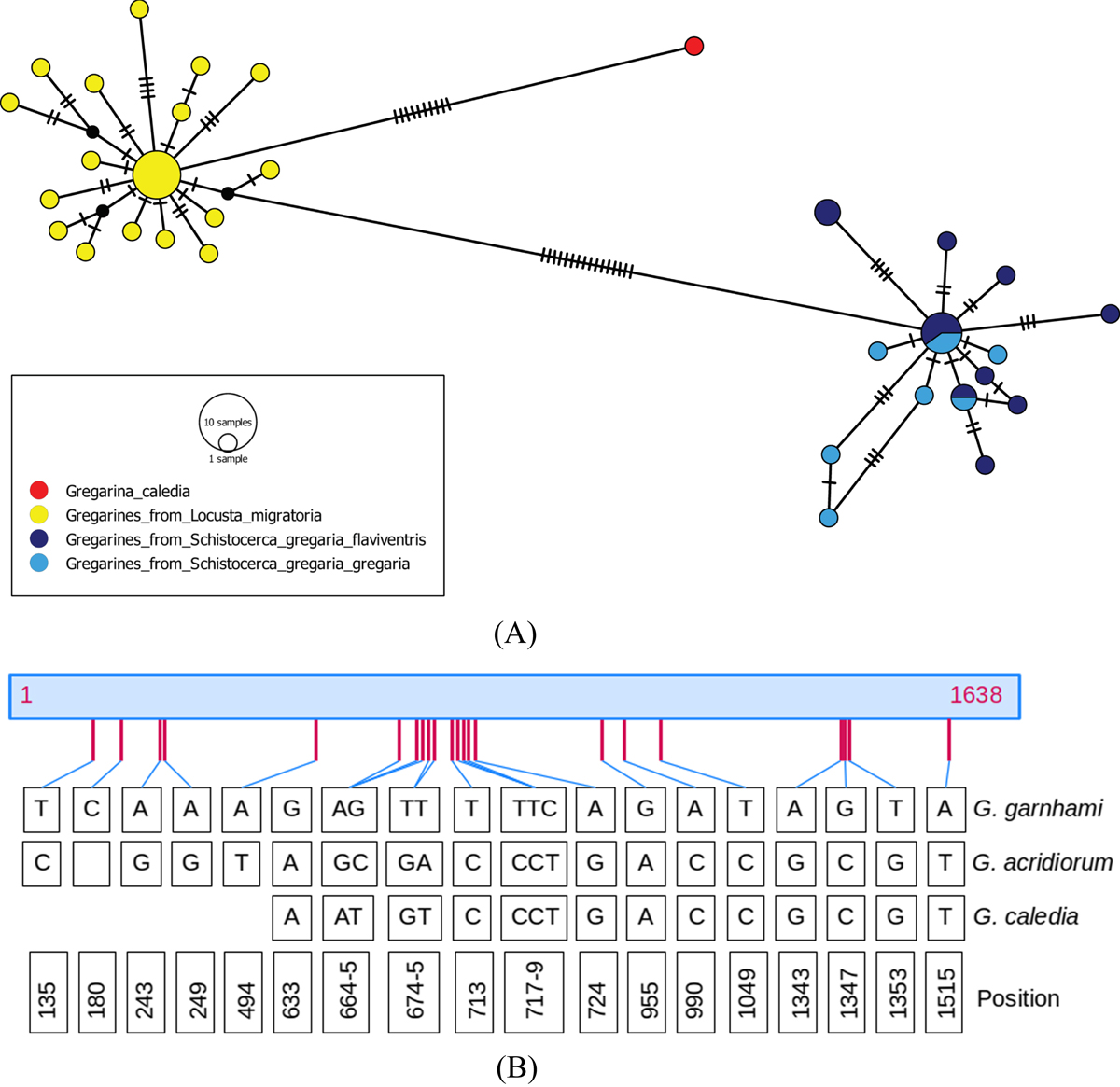
Download original image
A: Minimum spanning network for the 43 cloned sequences of the SSU rDNA region studied, and the published sequence of G. caledia (L31799). Each link between haplotypes indicates one mutation, including indel events. The colors indicate the species or subspecies of the host. This network was inferred using POPART [25]. B: Distribution of the 22 polymorphic positions in SSU rDNA locus regions V1-V8 (1638-bp), between type 1 (presumably G. garnhami) (n = 20) and type 2 (presumably G. acridiorum) (n = 23) sequences, amplified from gregarines parasitizing respectively S. gregaria and L. migratoria. The corresponding positions are also given for G. caledia (L31799, 1210 bp) parasitizing Caledia captiva. Eleven additional positions, otherwise conserved between G. garnhami and G. acridiorum sequences, are modified in G. caledia sequence: site 1059, G deletion; sites 1161-1164: GAGC substituted by AG-G; site 1181: G substituted for C; site 1187: G substituted for A; sites 1231 and 1240: T substituted for C; site 1493: T insertion; site 1584: G substituted for A.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


