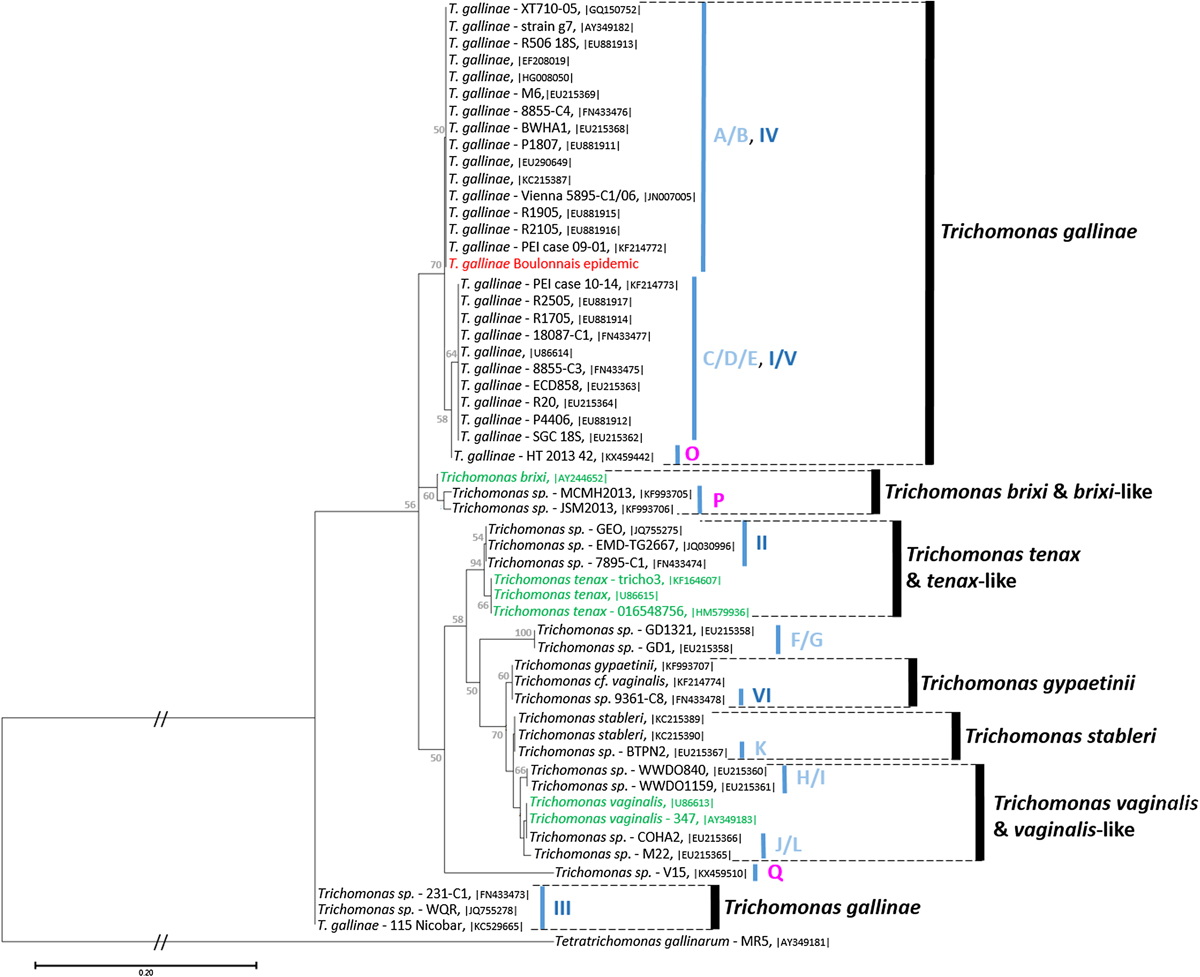
Figure 4
Download original image
Molecular phylogeny of avian Trichomonas and related species inferred on the internal transcribed spacer (ITS) region. The analysis was performed by Maximum Likelihood (ML) with the Tamura 3-parameter + Γ + I model of evolution [78] on a 371 bp fragment covering the ITS1–5.8SrRNA–ITS2 region of our sequences MK172843 in red, 47 sequences of avian Trichomonas in black, six related sequences of Trichomonas infecting mammals in green and Tetratrichomonas gallinarum as the outgroup to root the tree. Accession numbers of the sequences are provided between vertical bars. The reliability of tree topologies, nodal robustness, and statistical branch support were assessed by non-parametric bootstrap analysis using 1000 replicates (only >50% shown). Avian Trichomonas genotype names according to the classification of Gerhold et al. [29] and Grabsteinner et al. [34] are given in light and dark blue, respectively. Recent avian Trichomonas genotypes reported by Marx et al. [52] are given in purple.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


