Figure 2.
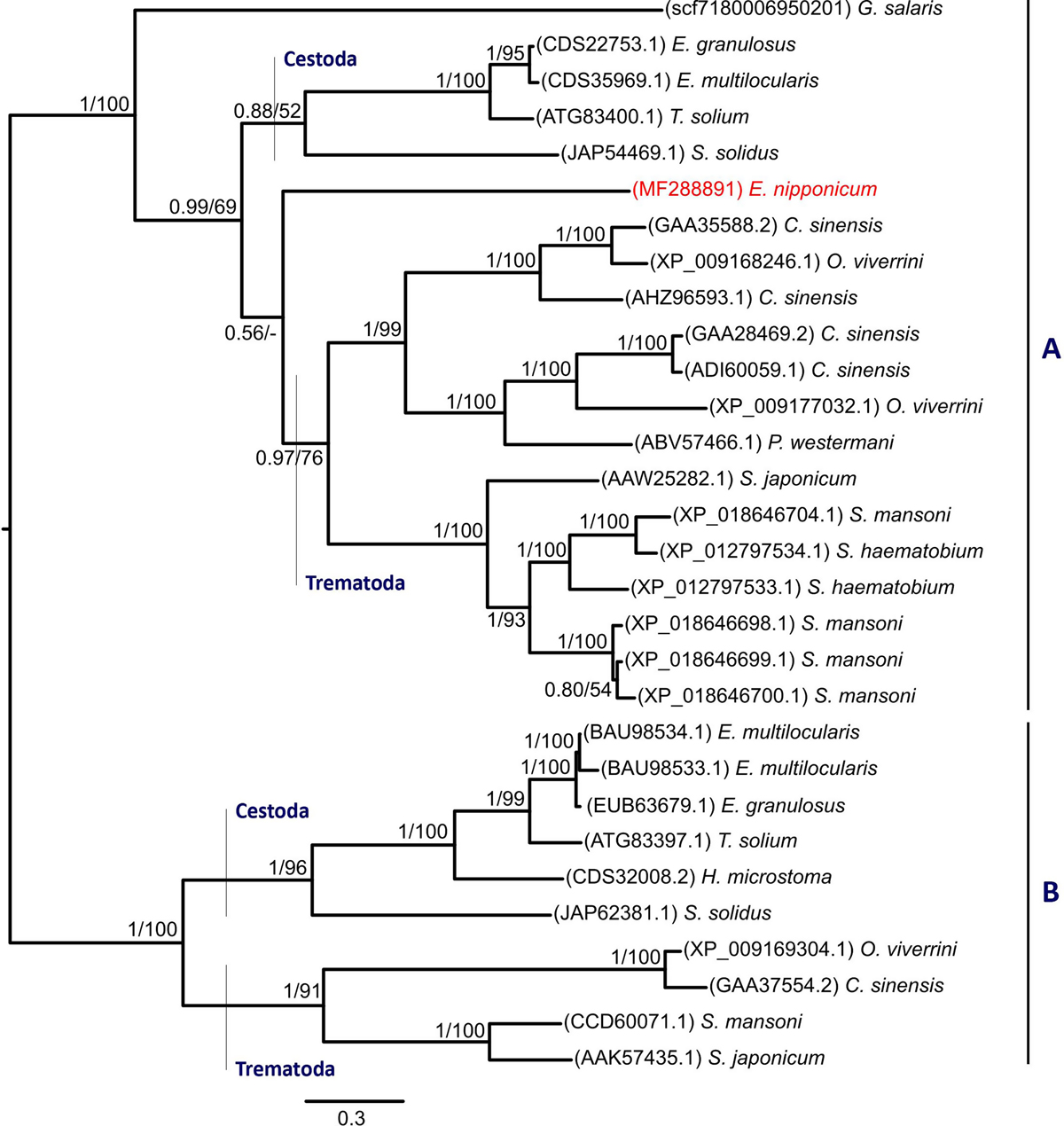
Download original image
A phylogram of platyhelminth serpin homologs, Bayesian inference analysis. Values along the branches indicate posterior probabilities and bootstrap values resulting from Bayesian inference and Maximum likelihood analyses, respectively. Proportional lengths of the branches correspond to the expected number of amino acid substitutions per site. The resulting tree is mid-point rooted in order to visualise the clustering of representative subfamilies. Newly obtained Eudiplozoon nipponicum serpin homolog (EnSerp1) is labelled red.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


