Figure 1.
Download original image
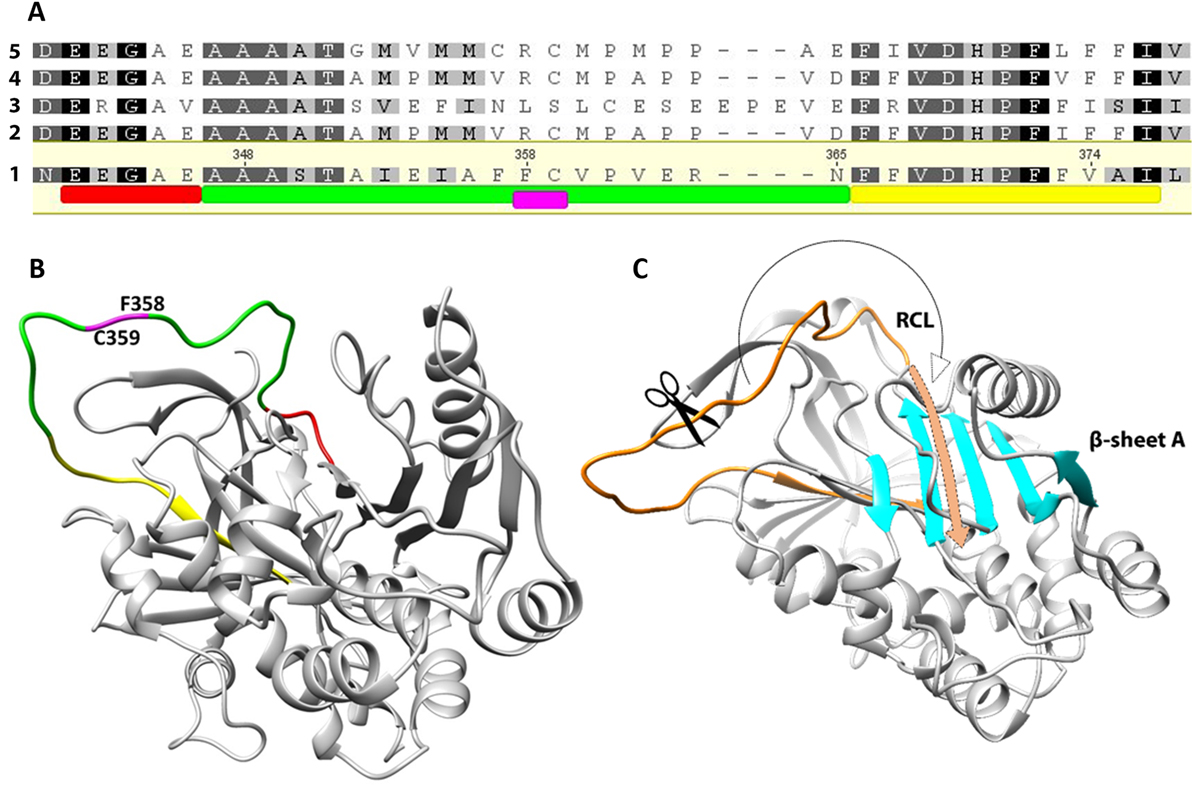
(A) Alignment of the EnSerp1 sequence with four of the most similar serpin sequences of other platyhelminths: 1, EnSerp1 from Eudiplozoon nipponicum (GenBank: MF288891.1); 2, Echinococcus multilocularis (GenBank: CDS35969.1); 3, Schistosoma haematobium (GenBank: XP_012797533.1); 4, Echinococcus granulosus (GenBank: CDS22753.1); 5, Taenia solium (GenBank: ATG83400.1). Conserved motifs characteristic for serpins are highlighted. The serpin motif (E342 – E346) shown in red is part of the reactive centre loop (RCL, A347 – N365), shown in green. Immediately after RCL, follows serpin signature (F366 – I376) in yellow. Scissile bond is situated within the RCL between P1 (F358) and P1’ (C359) residue, shown in magenta. (B) Predicted 3D structure of EnSerp1. Coloured areas of the molecule correspond to the sequence highlighted in Figure 1A. (C) RCL and β-sheet A. After the peptidase cleaves the scissile bond within RCL (in orange), the residual part of RCL is incorporated as a new strand into β-sheet A (in cyan).
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


