Figure 4
Download original image
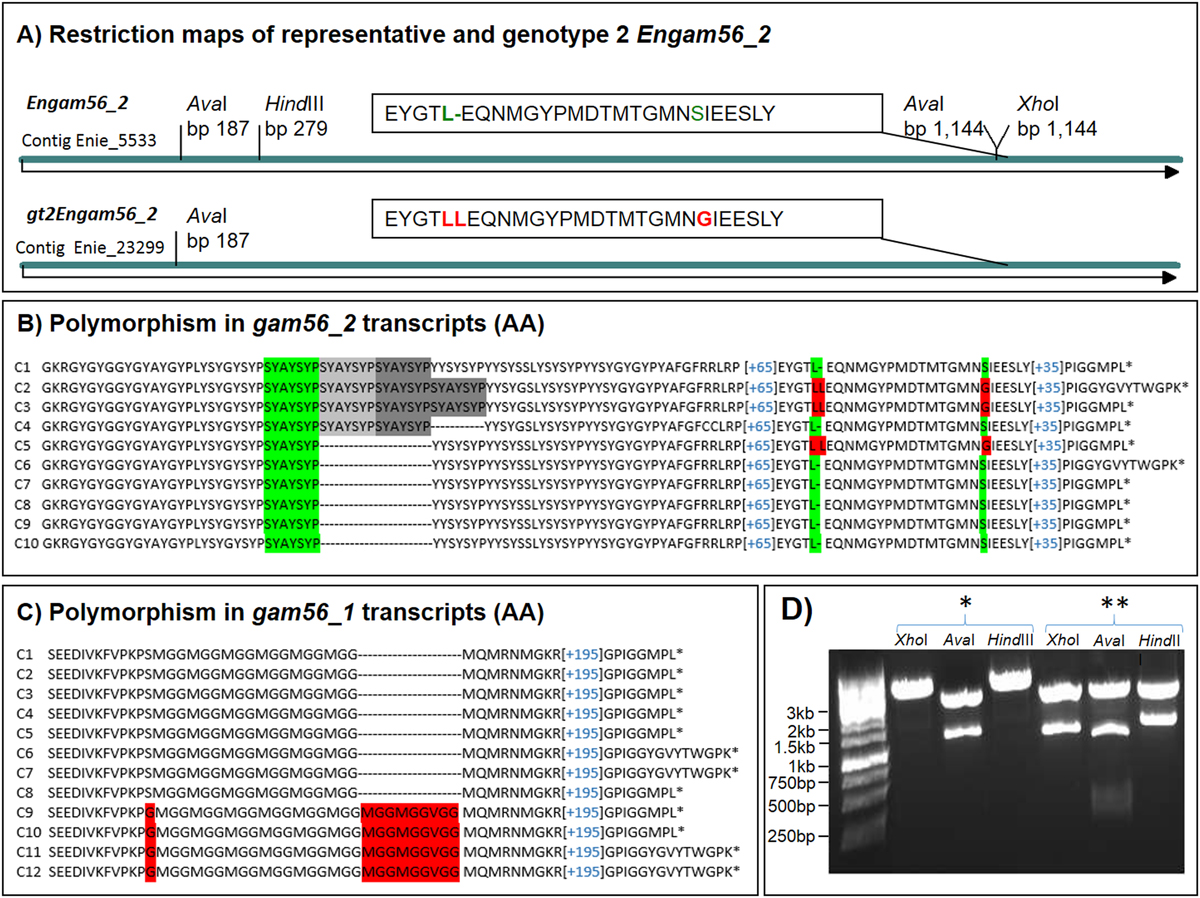
A) The major differences for identification of the gt2Engam56_2 (partially encoded by contig Enie_23299 and complete sequenced after PCR amplification) and the representative Engam56_2 (GenBank AJG00896.1) are different restriction sites (shown in D) and few amino acid changes (AA) (see A) LL(15 AA)G) vs. L(15 AA)S). For full DNA and translated sequences (AA) of the gt2EnGAM56_2, see SF1A and B.
B) Sequence analysis of Engam56_2 cDNA clones (translated sequence is shown) revealed a polymorph region (SYAYSYP motif) in some clones (C1-C4), which is present in both Engam56_2 versions. Both versions are also alternatively spliced.
C) Sequence analysis of Engam56_1 cDNA clones (translated sequence is shown) revealed two versions (red background, see also SF1D and E). C1-C8 belongs to the high-coverage gam locus assembly, whereas C9-C12 is associated with the low-coverage contig Enie_23088, which maps to contig Enie_5532. Please note that both versions were found spliced and unspliced.
D) The representative Engam56_2 and gt2Engam56_2 can be amplified by the same primer pair, but can be discriminated by restriction site analysis in plasmid DNA. The representative Engam56_2 (**) has two AvaI sites and harbours a HindIII and XhoI site, the gt2Engam56_2 (*) has only one AvaI site and no HindIII or XhoI site resulting in different fragment lengths in the agarose gel. The cloning vector pJET1.2 also contains these restriction sites at insert flanking positions.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


